Autophagy and aging
- PMID: 25916586
- PMCID: PMC4644734
- DOI: 10.1007/978-1-4939-2404-2_3
Autophagy and aging
Abstract
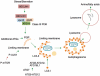
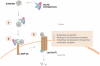
Autophagy is a critical quality control pathway that is conserved across diverse systems ranging from simple unicellular organisms like yeast to more complex systems, for instance mammals. Although, the fundamental role of autophagy is to maintain cellular quality control through lysosomal degradation of unwanted proteins and organelles, recent studies have mapped several new functions of this pathway that range from fuel utilization, cellular differentiation to protection against cell death. Given the importance of this pathway in maintaining cellular homeostasis, it has been considered that compromised autophagy could contribute to several of the commonly observed age-associated pathologies including neurodegeneration, reduction of muscle mass, cardiac malfunction, excessive lipid accumulation in tissues and glucose intolerance. The present chapter describes the two best-characterized autophagy pathways—macroautophagy and chaperone-mediated autophagy, and discusses how changes in these pathways associate with age-associated disorders. Understanding how to maintain “clean cells” by activation of autophagy could be an attractive strategy to maintain healthspan in aged individuals.
Figures
References
-
- Bergamini E, Del Roso A, Fierabracci V, Gori Z, Masiello P, Masini M, Pollera M. A new method for the investigation of endocrine-regulated autophagy and protein degradation in rat liver. Exp Mol Pathol. 1993;59(1):13–26. - PubMed
-
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW., 4th A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

