A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes
- PMID: 25916849
- PMCID: PMC4446428
- DOI: 10.1093/nar/gkv365
A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes
Abstract
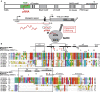
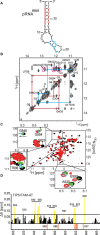
The chromatin remodeling complex NoRC, comprising the subunits SNF2h and TIP5/BAZ2A, mediates heterochromatin formation at major clusters of repetitive elements, including rRNA genes, centromeres and telomeres. Association with chromatin requires the interaction of the TAM (TIP5/ARBP/MBD) domain of TIP5 with noncoding RNA, which targets NoRC to specific genomic loci. Here, we show that the NMR structure of the TAM domain of TIP5 resembles the fold of the MBD domain, found in methyl-CpG binding proteins. However, the TAM domain exhibits an extended MBD fold with unique C-terminal extensions that constitute a novel surface for RNA binding. Mutation of critical amino acids within this surface abolishes RNA binding in vitro and in vivo. Our results explain the distinct binding specificities of TAM and MBD domains to RNA and methylated DNA, respectively, and reveal structural features for the interaction of NoRC with non-coding RNA.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- McStay B., Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. - PubMed
-
- Santoro R., Li J., Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

