New insights into the regulatory mechanisms of ppGpp and DksA on Escherichia coli RNA polymerase-promoter complex
- PMID: 25916853
- PMCID: PMC4446441
- DOI: 10.1093/nar/gkv391
New insights into the regulatory mechanisms of ppGpp and DksA on Escherichia coli RNA polymerase-promoter complex
Abstract
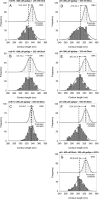
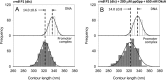
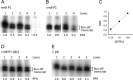
The stringent response modulators, guanosine tetraphosphate (ppGpp) and protein DksA, bind RNA polymerase (RNAP) and regulate gene expression to adapt bacteria to different environmental conditions. Here, we use Atomic Force Microscopy and in vitro transcription assays to study the effects of these modulators on the conformation and stability of the open promoter complex (RPo) formed at the rrnA P1, rrnB P1, its discriminator (dis) variant and λ pR promoters. In the absence of modulators, RPo formed at these promoters show different extents of DNA wrapping which correlate with the position of UP elements. Addition of the modulators affects both DNA wrapping and RPo stability in a promoter-dependent manner. Overall, the results obtained under different conditions of ppGpp, DksA and initiating nucleotides (iNTPs) indicate that ppGpp allosterically prevents the conformational changes associated with an extended DNA wrapping that leads to RPo stabilization, while DksA interferes directly with nucleotide positioning into the RNAP active site. At the iNTPs-sensitive rRNA promoters ppGpp and DksA display an independent inhibitory effect, while at the iNTPs-insensitive pR promoter DksA reduces the effect of ppGpp in accordance with their antagonistic role.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

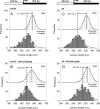
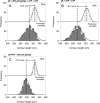
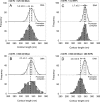
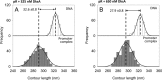
References
-
- Paul B.J., Barker M.M., Ross W., Schneider D.A., Webb C., Foster J.W., Gourse R.L. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. - PubMed
-
- Murray H.D., Schneider D.A., Gourse R.L. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell. 2003;12:125–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

