Exome sequencing reveals pathogenic mutations in 91 strains of mice with Mendelian disorders
- PMID: 25917818
- PMCID: PMC4484392
- DOI: 10.1101/gr.186882.114
Exome sequencing reveals pathogenic mutations in 91 strains of mice with Mendelian disorders
Abstract
Spontaneously arising mouse mutations have served as the foundation for understanding gene function for more than 100 years. We have used exome sequencing in an effort to identify the causative mutations for 172 distinct, spontaneously arising mouse models of Mendelian disorders, including a broad range of clinically relevant phenotypes. To analyze the resulting data, we developed an analytics pipeline that is optimized for mouse exome data and a variation database that allows for reproducible, user-defined data mining as well as nomination of mutation candidates through knowledge-based integration of sample and variant data. Using these new tools, putative pathogenic mutations were identified for 91 (53%) of the strains in our study. Despite the increased power offered by potentially unlimited pedigrees and controlled breeding, about half of our exome cases remained unsolved. Using a combination of manual analyses of exome alignments and whole-genome sequencing, we provide evidence that a large fraction of unsolved exome cases have underlying structural mutations. This result directly informs efforts to investigate the similar proportion of apparently Mendelian human phenotypes that are recalcitrant to exome sequencing.
© 2015 Fairfield et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
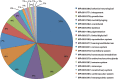
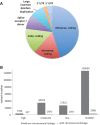

References
-
- Antonarakis SE, Beckmann JS. 2006. Mendelian disorders deserve more attention. Nat Rev Genet 7: 277–282. - PubMed
-
- Bailey DW. 1978. Sources of subline divergence and their relative importance for sublines of six major inbred strains of mice. Academic Press, Waltham, MA.
Publication types
MeSH terms
Grants and funding
- OD011163/OD/NIH HHS/United States
- R21 OD011163/OD/NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- HG006493/HG/NHGRI NIH HHS/United States
- EY015073/EY/NEI NIH HHS/United States
- U01 DE020052/DE/NIDCR NIH HHS/United States
- UM1 HG006493/HG/NHGRI NIH HHS/United States
- R01 EY019943/EY/NEI NIH HHS/United States
- OD010972/OD/NIH HHS/United States
- U54 HG006493/HG/NHGRI NIH HHS/United States
- R01EY019943/EY/NEI NIH HHS/United States
- R01 EY015073/EY/NEI NIH HHS/United States
- P40 OD010972/OD/NIH HHS/United States
- DE020052/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
