An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE
- PMID: 25918158
- PMCID: PMC4505474
- DOI: 10.1074/jbc.M115.650259
An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE
Abstract
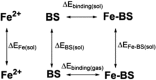
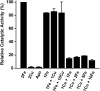
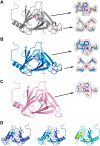
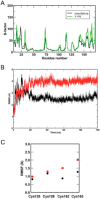
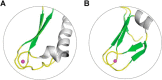
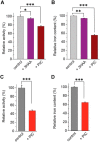
The rubredoxin motif is present in over 74,000 protein sequences and 2,000 structures, but few have known functions. A secondary, non-catalytic, rubredoxin-like iron site is conserved in 3-hydroxyanthranilate 3,4-dioxygenase (HAO), from single cellular sources but not multicellular sources. Through the population of the two metal binding sites with various metals in bacterial HAO, the structural and functional relationship of the rubredoxin-like site was investigated using kinetic, spectroscopic, crystallographic, and computational approaches. It is shown that the first metal presented preferentially binds to the catalytic site rather than the rubredoxin-like site, which selectively binds iron when the catalytic site is occupied. Furthermore, an iron ion bound to the rubredoxin-like site is readily delivered to an empty catalytic site of metal-free HAO via an intermolecular transfer mechanism. Through the use of metal analysis and catalytic activity measurements, we show that a downstream metabolic intermediate can selectively remove the catalytic iron. As the prokaryotic HAO is often crucial for cell survival, there is a need for ensuring its activity. These results suggest that the rubredoxin-like site is a possible auxiliary iron source to the catalytic center when it is lost during catalysis in a pathway with metabolic intermediates of metal-chelating properties. A spare tire concept is proposed based on this biochemical study, and this concept opens up a potentially new functional paradigm for iron-sulfur centers in iron-dependent enzymes as transient iron binding and shuttling sites to ensure full metal loading of the catalytic site.
Keywords: computation; dioxygenase; iron reservoir; iron-sulfur protein; kynurenine; metabolism; tryptophan.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




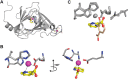
References
-
- Orme-Johnson W. H. (1973) Iron-sulfur proteins: structure and function. Annu. Rev. Biochem. 42, 159–204 - PubMed
-
- Adam V., Royant A., Nivière V., Molina-Heredia F. P., Bourgeois D. (2004) Structure of superoxide reductase bound to ferrocyanide and active site expansion upon x-ray-induced photo-reduction. Structure 12, 1729–1740 - PubMed
-
- Larsson K. M., Andersson J., Sjöberg B. M., Nordlund P., Logan D. T. (2001) Structural basis for allosteric substrate specificity regulation in anaerobic ribonucleotide reductases. Structure 9, 739–750 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

