Regulation of Structural Dynamics within a Signal Recognition Particle Promotes Binding of Protein Targeting Substrates
- PMID: 25918165
- PMCID: PMC4505461
- DOI: 10.1074/jbc.M114.624346
Regulation of Structural Dynamics within a Signal Recognition Particle Promotes Binding of Protein Targeting Substrates
Abstract
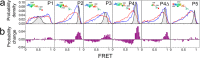
Protein targeting is critical in all living organisms and involves a signal recognition particle (SRP), an SRP receptor, and a translocase. In co-translational targeting, interactions among these proteins are mediated by the ribosome. In chloroplasts, the light-harvesting chlorophyll-binding protein (LHCP) in the thylakoid membrane is targeted post-translationally without a ribosome. A multidomain chloroplast-specific subunit of the SRP, cpSRP43, is proposed to take on the role of coordinating the sequence of targeting events. Here, we demonstrate that cpSRP43 exhibits significant interdomain dynamics that are reduced upon binding its SRP binding partner, cpSRP54. We showed that the affinity of cpSRP43 for the binding motif of LHCP (L18) increases when cpSRP43 is complexed to the binding motif of cpSRP54 (cpSRP54pep). These results support the conclusion that substrate binding to the chloroplast SRP is modulated by protein structural dynamics in which a major role of cpSRP54 is to improve substrate binding efficiency to the cpSRP.
Keywords: chloroplast; isothermal titration calorimetry (ITC); membrane transport; molecular dynamics; protein dynamic; protein targeting; signal recognition particle (SRP); single molecule biophysics.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Cline K. (2003) in Light-Harvesting Antennas in Photosynthesis (Green B., Parson W. W., eds) pp. 353–372, Kluwer Academic Publishers, Dordrecht, the Netherlands
-
- Henry R., Goforth R. L., Schunemann D. (2007) in The Enzymes (Dalbey R. E., Koehler C. M., Tamanoi F., eds), pp. 493–521, Academic Press, San Diego, CA
-
- Franklin A. E., Hoffman N. E. (1993) Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J. Biol. Chem. 268, 22175–22180 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

