Genomic Characterization of Non-Small-Cell Lung Cancer in African Americans by Targeted Massively Parallel Sequencing
- PMID: 25918285
- PMCID: PMC4451177
- DOI: 10.1200/JCO.2014.59.2444
Genomic Characterization of Non-Small-Cell Lung Cancer in African Americans by Targeted Massively Parallel Sequencing
Abstract
Purpose: Technologic advances have enabled the comprehensive analysis of genetic perturbations in non-small-cell lung cancer (NSCLC); however, African Americans have often been underrepresented in these studies. This ethnic group has higher lung cancer incidence and mortality rates, and some studies have suggested a lower incidence of epidermal growth factor receptor mutations. Herein, we report the most in-depth molecular profile of NSCLC in African Americans to date.
Methods: A custom panel was designed to cover the coding regions of 81 NSCLC-related genes and 40 ancestry-informative markers. Clinical samples were sequenced on a massively parallel sequencing instrument, and anaplastic lymphoma kinase translocation was evaluated by fluorescent in situ hybridization.
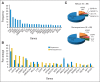
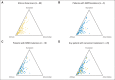
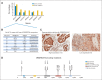
Results: The study cohort included 99 patients (61% males, 94% smokers) comprising 31 squamous and 68 nonsquamous cell carcinomas. We detected 227 nonsilent variants in the coding sequence, including 24 samples with nonoverlapping, classic driver alterations. The frequency of driver mutations was not significantly different from that of whites, and no association was found between genetic ancestry and the presence of somatic mutations. Copy number alteration analysis disclosed distinguishable amplifications in the 3q chromosome arm in squamous cell carcinomas and pointed toward a handful of targetable alterations. We also found frequent SMARCA4 mutations and protein loss, mostly in driver-negative tumors.
Conclusion: Our data suggest that African American ancestry may not be significantly different from European/white background for the presence of somatic driver mutations in NSCLC. Furthermore, we demonstrated that using a comprehensive genotyping approach could identify numerous targetable alterations, with potential impact on therapeutic decisions.
© 2015 by American Society of Clinical Oncology.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest are found in the article online at
Figures


Comment in
-
Are there any differences in genomic characterization of non-small cell lung cancer between African Americans and Whites?J Thorac Dis. 2016 Nov;8(11):E1517-E1519. doi: 10.21037/jtd.2016.11.65. J Thorac Dis. 2016. PMID: 28066648 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

