Modular expression analysis reveals functional conservation between human Langerhans cells and mouse cross-priming dendritic cells
- PMID: 25918340
- PMCID: PMC4419344
- DOI: 10.1084/jem.20131675
Modular expression analysis reveals functional conservation between human Langerhans cells and mouse cross-priming dendritic cells
Abstract
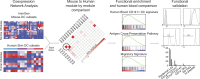
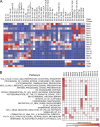
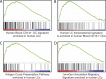
Characterization of functionally distinct dendritic cell (DC) subsets in mice has fueled interest in whether analogous counterparts exist in humans. Transcriptional modules of coordinately expressed genes were used for defining shared functions between the species. Comparing modules derived from four human skin DC subsets and modules derived from the Immunological Genome Project database for all mouse DC subsets revealed that human Langerhans cells (LCs) and the mouse XCR1(+)CD8α(+)CD103(+) DCs shared the class I-mediated antigen processing and cross-presentation transcriptional modules that were not seen in mouse LCs. Furthermore, human LCs were enriched in a transcriptional signature specific to the blood cross-presenting CD141/BDCA-3(+) DCs, the proposed equivalent to mouse CD8α(+) DCs. Consistent with our analysis, LCs were highly adept at inducing primary CTL responses. Thus, our study suggests that the function of LCs may not be conserved between mouse and human and supports human LCs as an especially relevant therapeutic target.
© 2015 Artyomov et al.
Figures




References
-
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. . 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281 10.1084/jem.20100348 - DOI - PMC - PubMed
-
- Banchereau J., Thompson-Snipes L., Zurawski S., Blanck J.P., Cao Y., Clayton S., Gorvel J.P., Zurawski G., and Klechevsky E.. 2012a. The differential production of cytokines by human Langerhans cells and dermal CD14(+) DCs controls CTL priming. Blood. 119:5742–5749 10.1182/blood-2011-08-371245 - DOI - PMC - PubMed
-
- Banchereau J., Zurawski S., Thompson-Snipes L., Blanck J.P., Clayton S., Munk A., Cao Y., Wang Z., Khandelwal S., Hu J., et al. . 2012b. Immunoglobulin-like transcript receptors on human dermal CD14+ dendritic cells act as a CD8-antagonist to control cytotoxic T cell priming. Proc. Natl. Acad. Sci. USA. 109:18885–18890 10.1073/pnas.1205785109 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

