Specific bone cells produce DLL4 to generate thymus-seeding progenitors from bone marrow
- PMID: 25918341
- PMCID: PMC4419348
- DOI: 10.1084/jem.20141843
Specific bone cells produce DLL4 to generate thymus-seeding progenitors from bone marrow
Abstract
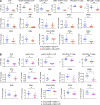
Production of the cells that ultimately populate the thymus to generate α/β T cells has been controversial, and their molecular drivers remain undefined. Here, we report that specific deletion of bone-producing osteocalcin (Ocn)-expressing cells in vivo markedly reduces T-competent progenitors and thymus-homing receptor expression among bone marrow hematopoietic cells. Decreased intrathymic T cell precursors and decreased generation of mature T cells occurred despite normal thymic function. The Notch ligand DLL4 is abundantly expressed on bone marrow Ocn(+) cells, and selective depletion of DLL4 from these cells recapitulated the thymopoietic abnormality. These data indicate that specific mesenchymal cells in bone marrow provide key molecular drivers enforcing thymus-seeding progenitor generation and thereby directly link skeletal biology to the production of T cell-based adaptive immunity.
© 2015 Yu et al.
Figures









Comment in
-
T cell development runs marrow deep.J Exp Med. 2015 May 4;212(5):599-600. doi: 10.1084/jem.2125insight3. J Exp Med. 2015. PMID: 25941319 Free PMC article. No abstract available.
References
-
- Awong G., Singh J., Mohtashami M., Malm M., La Motte-Mohs R.N., Benveniste P.M., Serra P., Herer E., van den Brink M.R., and Zúñiga-Pflücker J.C.. 2013. Human proT-cells generated in vitro facilitate hematopoietic stem cell-derived T-lymphopoiesis in vivo and restore thymic architecture. Blood. 122:4210–4219 10.1182/blood-2012-12-472803 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

