How release of phosphate from mammalian F1-ATPase generates a rotary substep
- PMID: 25918412
- PMCID: PMC4434703
- DOI: 10.1073/pnas.1506465112
How release of phosphate from mammalian F1-ATPase generates a rotary substep
Abstract
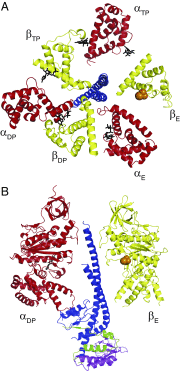
The rotation of the central stalk of F1-ATPase is driven by energy derived from the sequential binding of an ATP molecule to its three catalytic sites and the release of the products of hydrolysis. In human F1-ATPase, each 360° rotation consists of three 120° steps composed of substeps of about 65°, 25°, and 30°, with intervening ATP binding, phosphate release, and catalytic dwells, respectively. The F1-ATPase inhibitor protein, IF1, halts the rotary cycle at the catalytic dwell. The human and bovine enzymes are essentially identical, and the structure of bovine F1-ATPase inhibited by IF1 represents the catalytic dwell state. Another structure, described here, of bovine F1-ATPase inhibited by an ATP analog and the phosphate analog, thiophosphate, represents the phosphate binding dwell. Thiophosphate is bound to a site in the α(E)β(E)-catalytic interface, whereas in F1-ATPase inhibited with IF1, the equivalent site is changed subtly and the enzyme is incapable of binding thiophosphate. These two structures provide a molecular mechanism of how phosphate release generates a rotary substep as follows. In the active enzyme, phosphate release from the β(E)-subunit is accompanied by a rearrangement of the structure of its binding site that prevents released phosphate from rebinding. The associated extrusion of a loop in the β(E)-subunit disrupts interactions in the α(E)β(E-)catalytic interface and opens it to its fullest extent. Other rearrangements disrupt interactions between the γ-subunit and the C-terminal domain of the α(E)-subunit. To restore most of these interactions, and to make compensatory new ones, the γ-subunit rotates through 25°-30°.
Keywords: ATP synthase; mitochondria; phosphate release; rotary substep.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Walker JE, Lutter R, Dupuis A, Runswick MJ. Identification of the subunits of F1Fo-ATPase from bovine heart mitochondria. Biochemistry. 1991;30(22):5369–5378. - PubMed
-
- Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41(1):1–16. - PubMed
-
- Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

