The human gut resistome
- PMID: 25918444
- PMCID: PMC4424436
- DOI: 10.1098/rstb.2014.0087
The human gut resistome
Abstract
In recent decades, the emergence and spread of antibiotic resistance among bacterial pathogens has become a major threat to public health. Bacteria can acquire antibiotic resistance genes by the mobilization and transfer of resistance genes from a donor strain. The human gut contains a densely populated microbial ecosystem, termed the gut microbiota, which offers ample opportunities for the horizontal transfer of genetic material, including antibiotic resistance genes. Recent technological advances allow microbiota-wide studies into the diversity and dynamics of the antibiotic resistance genes that are harboured by the gut microbiota ('the gut resistome'). Genes conferring resistance to antibiotics are ubiquitously present among the gut microbiota of humans and most resistance genes are harboured by strictly anaerobic gut commensals. The horizontal transfer of genetic material, including antibiotic resistance genes, through conjugation and transduction is a frequent event in the gut microbiota, but mostly involves non-pathogenic gut commensals as these dominate the microbiota of healthy individuals. Resistance gene transfer from commensals to gut-dwelling opportunistic pathogens appears to be a relatively rare event but may contribute to the emergence of multi-drug resistant strains, as is illustrated by the vancomycin resistance determinants that are shared by anaerobic gut commensals and the nosocomial pathogen Enterococcus faecium.
Keywords: Enterococcus faecium; antibiotic resistance; gut commensals; gut microbiota; metagenomics.
Figures
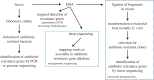

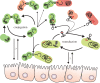
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

