Leprous lesion presents enrichment of opportunistic pathogenic bacteria
- PMID: 25918684
- PMCID: PMC4405507
- DOI: 10.1186/s40064-015-0955-1
Leprous lesion presents enrichment of opportunistic pathogenic bacteria
Abstract
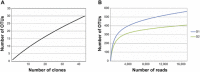
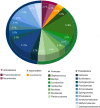
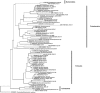
Leprosy is a chronic infectious disease that remains a major challenge to public health in endemic countries. Increasing evidence has highlighted the importance of microbiota for human general health and, as such, the study of skin microbiota is of interest. But while studies are continuously revealing the complexity of human skin microbiota, the microbiota of leprous cutaneous lesions has not yet been characterized. Here we used Sanger and massively parallel small sub-unit rRNA (SSU) rRNA gene sequencing to characterize the microbiota of leprous lesions, and studied how it differs from the bacterial skin composition of healthy individuals previously described in the literature. Taxonomic analysis of leprous lesions revealed main four phyla: Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, with Proteobacteria presenting the highest diversity. There were considerable differences in the distribution of Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria, with the first two phyla enriched and the other markedly diminished in the leprous lesions, when compared with healthy skin. Propionibacterium, Corynebacterium and Staphylococcus, resident and abundant in healthy skin, were underrepresented in skin from leprous lesions. Most of the taxa found in skin from leprous lesions are not typical in human skin and potentially pathogenic, with the Burkholderia, Pseudomonas and Bacillus genera being overrepresented. Our data suggest significant shifts of the microbiota with emergence and competitive advantage of potentially pathogenic bacteria over skin resident taxa.
Keywords: 16S rRNA gene; Diversity; Leprosy; Microbiota; Skin.
Figures

References
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

