Metagenomic and lipid analyses reveal a diel cycle in a hypersaline microbial ecosystem
- PMID: 25918833
- PMCID: PMC4817636
- DOI: 10.1038/ismej.2015.66
Metagenomic and lipid analyses reveal a diel cycle in a hypersaline microbial ecosystem
Abstract
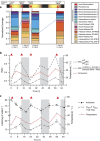
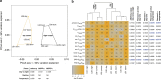
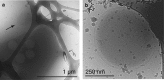
Marine microbial communities experience daily fluctuations in light and temperature that can have important ramifications for carbon and nutrient cycling. Elucidation of such short time scale community-wide dynamics is hindered by system complexity. Hypersaline aquatic environments have lower species richness than marine environments and can be well-defined spatially, hence they provide a model system for diel cycle analysis. We conducted a 3-day time series experiment in a well-defined pool in hypersaline Lake Tyrrell, Australia. Microbial communities were tracked by combining cultivation-independent lipidomic, metagenomic and microscopy methods. The ratio of total bacterial to archaeal core lipids in the planktonic community increased by up to 58% during daylight hours and decreased by up to 32% overnight. However, total organism abundances remained relatively consistent over 3 days. Metagenomic analysis of the planktonic community composition, resolved at the genome level, showed dominance by Haloquadratum species and six uncultured members of the Halobacteriaceae. The post 0.8 μm filtrate contained six different nanohaloarchaeal types, three of which have not been identified previously, and cryo-transmission electron microscopy imaging confirmed the presence of small cells. Notably, these nano-sized archaea showed a strong diel cycle, with a pronounced increase in relative abundance over the night periods. We detected no eukaryotic algae or other photosynthetic primary producers, suggesting that carbon resources may derive from patchily distributed microbial mats at the sediment-water interface or from surrounding land. Results show the operation of a strong community-level diel cycle, probably driven by interconnected temperature, light abundance, dissolved oxygen concentration and nutrient flux effects.
Figures




References
-
- Allen EE, Banfield JF. (2005). Community genomics in microbial ecology and evolution. Nat Rev Microbiol 3: 489–498. - PubMed
-
- Baker BJ, Banfield JF. (2003). Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44: 139–152. - PubMed
-
- Baliga NS. (2008). Systems biology - the scale of prediction. Science 320: 1297–1298. - PubMed
-
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate — a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300.
-
- Butte W. (1983). Rapid method for the determination of fatty-acid profiles from fats and oils using trimethylsulfonium hydroxiden for trans-esterification. J Chromatogr 261: 142–145.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

