The protein quality control machinery regulates its misassembled proteasome subunits
- PMID: 25919710
- PMCID: PMC4412499
- DOI: 10.1371/journal.pgen.1005178
The protein quality control machinery regulates its misassembled proteasome subunits
Abstract
Cellular toxicity introduced by protein misfolding threatens cell fitness and viability. Failure to eliminate these polypeptides is associated with various aggregation diseases. In eukaryotes, the ubiquitin proteasome system (UPS) plays a vital role in protein quality control (PQC), by selectively targeting misfolded proteins for degradation. While the assembly of the proteasome can be naturally impaired by many factors, the regulatory pathways that mediate the sorting and elimination of misassembled proteasomal subunits are poorly understood. Here, we reveal how the dysfunctional proteasome is controlled by the PQC machinery. We found that among the multilayered quality control mechanisms, UPS mediated degradation of its own misassembled subunits is the favored pathway. We also demonstrated that the Hsp42 chaperone mediates an alternative pathway, the accumulation of these subunits in cytoprotective compartments. Thus, we show that proteasome homeostasis is controlled through probing the level of proteasome assembly, and the interplay between UPS mediated degradation or their sorting into distinct cellular compartments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

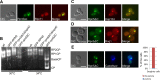
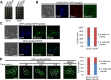

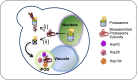
Similar articles
-
Spatial Organization of Proteasome Aggregates in the Regulation of Proteasome Homeostasis.Front Mol Biosci. 2020 Jan 9;6:150. doi: 10.3389/fmolb.2019.00150. eCollection 2019. Front Mol Biosci. 2020. PMID: 31998748 Free PMC article. Review.
-
The extent of Ssa1/Ssa2 Hsp70 chaperone involvement in nuclear protein quality control degradation varies with the substrate.Mol Biol Cell. 2020 Feb 1;31(3):221-233. doi: 10.1091/mbc.E18-02-0121. Epub 2019 Dec 11. Mol Biol Cell. 2020. PMID: 31825716 Free PMC article.
-
Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting.Mol Biol Cell. 2013 Jul;24(13):2076-87. doi: 10.1091/mbc.E13-01-0010. Epub 2013 May 1. Mol Biol Cell. 2013. PMID: 23637465 Free PMC article.
-
Hsp70-Hsp110 chaperones deliver ubiquitin-dependent and -independent substrates to the 26S proteasome for proteolysis in yeast.J Cell Sci. 2018 Mar 20;131(6):jcs210948. doi: 10.1242/jcs.210948. J Cell Sci. 2018. PMID: 29507114
-
Nuclear Ubiquitin-Proteasome Pathways in Proteostasis Maintenance.Biomolecules. 2021 Jan 4;11(1):54. doi: 10.3390/biom11010054. Biomolecules. 2021. PMID: 33406777 Free PMC article. Review.
Cited by
-
Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer.Nat Commun. 2018 Nov 16;9(1):4700. doi: 10.1038/s41467-018-07178-x. Nat Commun. 2018. PMID: 30446660 Free PMC article.
-
Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation.Elife. 2018 Apr 6;7:e34532. doi: 10.7554/eLife.34532. Elife. 2018. Retraction in: Elife. 2022 Sep 02;11:e82988. doi: 10.7554/eLife.82988. PMID: 29624167 Free PMC article. Retracted.
-
Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast.Mol Biol Cell. 2017 Sep 15;28(19):2479-2491. doi: 10.1091/mbc.E17-03-0162. Epub 2017 Aug 2. Mol Biol Cell. 2017. PMID: 28768827 Free PMC article.
-
Role of sHsps in organizing cytosolic protein aggregation and disaggregation.Cell Stress Chaperones. 2017 Jul;22(4):493-502. doi: 10.1007/s12192-017-0762-4. Epub 2017 Jan 24. Cell Stress Chaperones. 2017. PMID: 28120291 Free PMC article. Review.
-
Quantitative time-resolved analysis reveals intricate, differential regulation of standard- and immuno-proteasomes.Elife. 2015 Sep 22;4:e07545. doi: 10.7554/eLife.07545. Elife. 2015. PMID: 26393687 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

