Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity
- PMID: 25919952
- PMCID: PMC4408529
- DOI: 10.7554/eLife.06416
Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity
Abstract
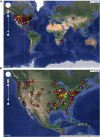
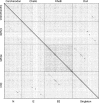
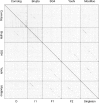
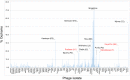
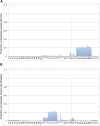
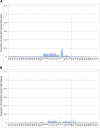
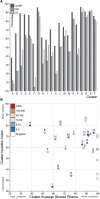
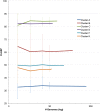
The bacteriophage population is large, dynamic, ancient, and genetically diverse. Limited genomic information shows that phage genomes are mosaic, and the genetic architecture of phage populations remains ill-defined. To understand the population structure of phages infecting a single host strain, we isolated, sequenced, and compared 627 phages of Mycobacterium smegmatis. Their genetic diversity is considerable, and there are 28 distinct genomic types (clusters) with related nucleotide sequences. However, amino acid sequence comparisons show pervasive genomic mosaicism, and quantification of inter-cluster and intra-cluster relatedness reveals a continuum of genetic diversity, albeit with uneven representation of different phages. Furthermore, rarefaction analysis shows that the mycobacteriophage population is not closed, and there is a constant influx of genes from other sources. Phage isolation and analysis was performed by a large consortium of academic institutions, illustrating the substantial benefits of a disseminated, structured program involving large numbers of freshman undergraduates in scientific discovery.
Keywords: bacteriophage; evolution; evolutionary biology; genomics; infectious disease; microbiology; viruses.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
-
- Cresawn SG, Pope WH, Jacobs-Sera D, Bowman CA, Russell DA, Dedrick RA, Adair T, Anders KR, Ball S, Bollivar D, Breitenberger C, Burnett SH, Butela K, Byrnes D, Carzo S, Cornely KA, Cross T, Daniels RL, Dunbar D, Findley AM, Gissendanner CR, Golebiewska UP, Hartzog GA, Hatherill JR, Hughes LE, Jalloh CS, De Los Santos C, Ekanam K, Khambule SL, King RA, King-Smith C, Klyczek K, Krukonis GP, Laing C, Lapin JS, Lopez AJ, Mkhwanazi SM, Molloy SD, Moran D, Munsamy V, Pacey E, Plymale R, Poxleitner M, Reyna N, Schildbach JF, Stukey J, Taylor SE, Ware VC, Wellmann AL, Westholm D, Wodarski D, Zajko M, Zikalala TS, Hendrix RW, Hatfull GF. Comparative genomics of cluster O mycobacteriophages. PLOS ONE. 2015;10:e0118725. doi: 10.1371/journal.pone.0118725. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM103430/GM/NIGMS NIH HHS/United States
- SC1 AI098976/AI/NIAID NIH HHS/United States
- GM51975/GM/NIGMS NIH HHS/United States
- GM103408/GM/NIGMS NIH HHS/United States
- R15 GM110578/GM/NIGMS NIH HHS/United States
- R01 GM051975/GM/NIGMS NIH HHS/United States
- P20 GM103430/GM/NIGMS NIH HHS/United States
- P20 GM103408/GM/NIGMS NIH HHS/United States
- GM1003423/GM/NIGMS NIH HHS/United States
- T32 GM008490/GM/NIGMS NIH HHS/United States
- 52007054/Howard Hughes Medical Institute/United States
- 54308198/Howard Hughes Medical Institute/United States
- GM094712/GM/NIGMS NIH HHS/United States
- P20 GM103423/GM/NIGMS NIH HHS/United States
- R25 GM090084/GM/NIGMS NIH HHS/United States
- R15 GM094712/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

