Docosahexaenoic acid differentially affects TNFα and IL-6 expression in LPS-stimulated RAW 264.7 murine macrophages
- PMID: 25921297
- PMCID: PMC4562472
- DOI: 10.1016/j.plefa.2015.03.002
Docosahexaenoic acid differentially affects TNFα and IL-6 expression in LPS-stimulated RAW 264.7 murine macrophages
Abstract
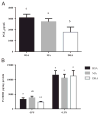
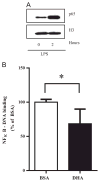
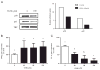
Docosahexaenoic acid (DHA) is generally reported to have anti-inflammatory properties, however, prior work has documented differential effects on individual pro-inflammatory cytokines: reduced IL-6, but not TNFα, mRNA expression in macrophages. To elucidate the mechanism, the roles of prostaglandin E2 (PGE2), cyclic AMP response element-binding protein (CREB), and NFκB were examined in RAW 264.7 macrophages. DHA did not influence CREB activity, but significantly reduced PGE2 production by 41% and NFκB activity by 32%. Exogenous PGE2 inhibited TNFα mRNA expression dose dependently. Unexpectedly, inhibiting PGE2 production with NS-398 also decreased TNFα mRNA expression, suggesting a concentration-dependent dual role of PGE2 in regulating TNFα expression. IL-6 expression was unaffected by endogenous or exogenous PGE2. Partial block of NFκB activation (SN50; 46%, or, BAY-11-7082; 41%) lowered IL-6 to a greater extent than TNFα mRNA expression. The differential effect of DHA on TNFα and IL-6 mRNA expression may be mediated via reduction in NFκB activity.
Keywords: CREB; IL-6; Macrophages; PGE(2); TLR4; TNFα.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
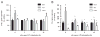



References
-
- Calder PC. Long-chain fatty acids and inflammation. Proc Nutr Soc. 2012;71:284–289. - PubMed
-
- Calder PC, Ahluwalia N, Albers R, et al. A consideration of biomarkers to be used for evaluation of inflammation in human nutritional studies. Br J Nutr. 2013;109(Suppl 1):S1–S34. - PubMed
-
- Branen L, Hovgaard L, Nitulescu M, Bengtsson E, Nilsson J, Jovinge S. Inhibition of tumor necrosis factor-alpha reduces atherosclerosis in apolipo-protein E knockout mice. Arterioscler Thromb Vasc Biol. 2004;24:2137–2142. - PubMed
-
- Sun Y, Yin M, Zhang L, Pan J. Characterization of the cytokine expression profiles of the aorta and liver of young tumor necrosis factor alpha mutant mice. Mol Cell Biochem. 2012;366:59–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

