DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function
- PMID: 25921537
- PMCID: PMC4426031
- DOI: 10.1016/j.celrep.2015.03.069
DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function
Abstract
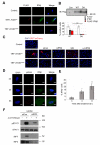
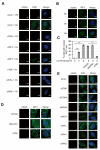
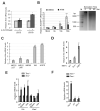
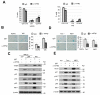
Expression of type I interferons (IFNs) can be induced by DNA-damaging agents, but the mechanisms and significance of this regulation are not completely understood. We found that the transcription factor IRF3, activated in an ATM-IKKα/β-dependent manner, stimulates cell-autonomous IFN-β expression in response to double-stranded DNA breaks. Cells and tissues with accumulating DNA damage produce endogenous IFN-β and stimulate IFN signaling in vitro and in vivo. In turn, IFN acts to amplify DNA-damage responses, activate the p53 pathway, promote senescence, and inhibit stem cell function in response to telomere shortening. Inactivation of the IFN pathway abrogates the development of diverse progeric phenotypes and extends the lifespan of Terc knockout mice. These data identify DNA-damage-response-induced IFN signaling as a critical mechanism that links accumulating DNA damage with senescence and premature aging.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev. 2011a;243:99–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

