The Mechanism and Function of Epigenetics in Uterine Leiomyoma Development
- PMID: 25922306
- PMCID: PMC5933172
- DOI: 10.1177/1933719115584449
The Mechanism and Function of Epigenetics in Uterine Leiomyoma Development
Abstract
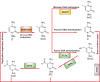
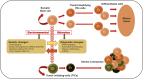
Uterine leiomyomas, also known as uterine fibroids, are the most common pelvic tumors, occurring in nearly 70% of all reproductive-aged women and are the leading indication for hysterectomy worldwide. The development of uterine leiomyomas involve a complex and heterogeneous constellation of hormones, growth factors, stem cells, genetic, and epigenetic abnormalities. An increasing body of evidence emphasizes the important contribution of epigenetics in the pathogenesis of leiomyomas. Genome-wide methylation analysis demonstrates that a subset of estrogen receptor (ER) response genes exhibit abnormal hypermethylation levels that are inversely correlated with their RNA expression. Several tumor suppressor genes, including Kruppel-like factor 11 (KLF11), deleted in lung and esophageal cancer 1 (DLEC1), keratin 19 (KRT19), and death-associated protein kinase 1 (DAPK1) also display higher hypermethylation levels in leiomyomas when compared to adjacent normal tissues. The important role of active DNA demethylation was recently identified with regard to the ten-eleven translocation protein 1 and ten-eleven translocation protein 3-mediated elevated levels of 5-hydroxymethylcytosine in leiomyoma. In addition, both histone deacetylase and histone methyltransferase are reported to be involved in the biology of leiomyomas. A number of deregulated microRNAs have been identified in leiomyomas, leading to an altered expression of their targets. More recently, the existence of side population (SP) cells with characteristics of tumor-initiating cells have been characterized in leiomyomas. These SP cells exhibit a tumorigenic capacity in immunodeficient mice when exposed to 17β-estradiol and progesterone, giving rise to fibroid-like tissue in vivo. These new findings will likely enhance our understanding of the crucial role epigenetics plays in the pathogenesis of uterine leiomyomas as well as point the way to novel therapeutic options.
Keywords: DNA methylation; TET proteins; epigenetics; histone modification; leiomyoma; miRNA; oxidative DNA demethylation; stem cells.
© The Author(s) 2015.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

