Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria
- PMID: 25922666
- PMCID: PMC4411801
- DOI: 10.1186/s40168-015-0077-6
Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria
Abstract
Background: Estuaries are among the most productive habitats on the planet. Bacteria in estuary sediments control the turnover of organic carbon and the cycling of nitrogen and sulfur. These communities are complex and primarily made up of uncultured lineages, thus little is known about how ecological and metabolic processes are partitioned in sediments.
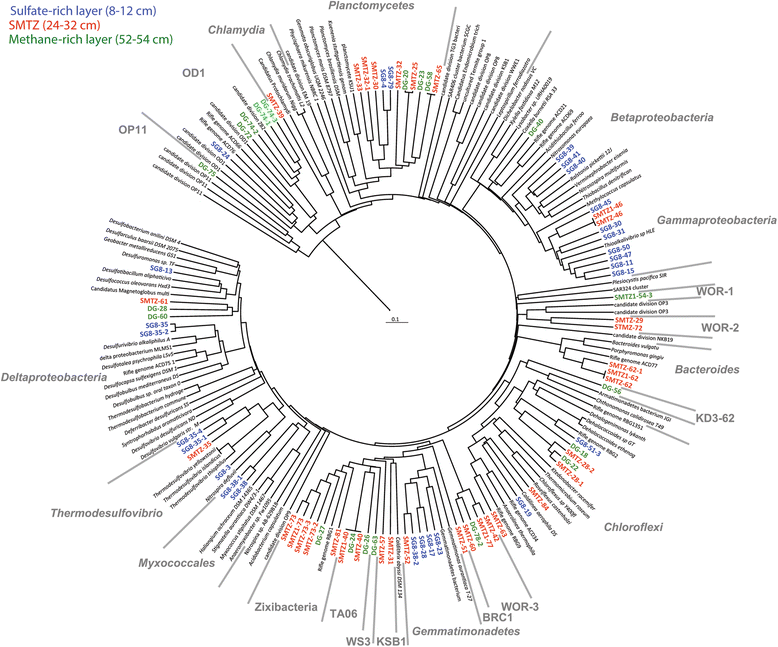
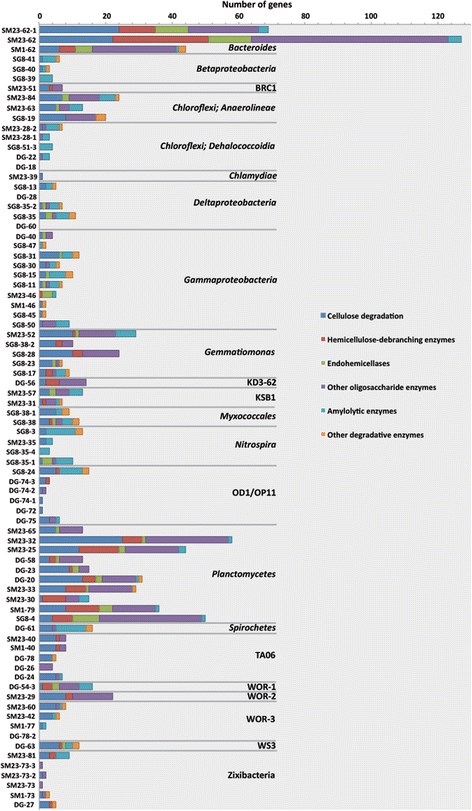
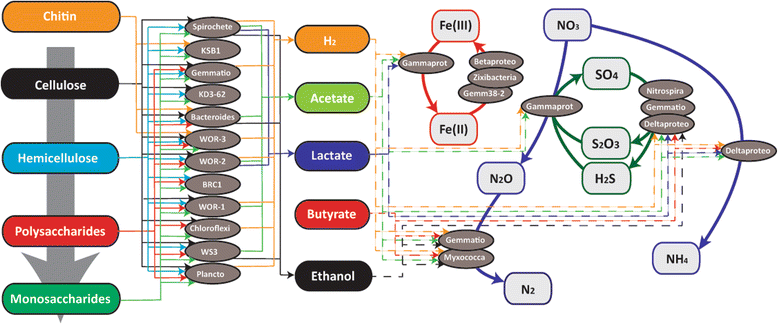
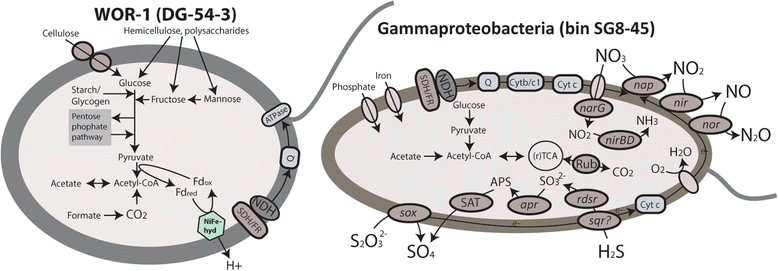
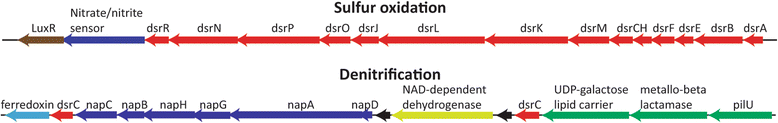
Results: De novo assembly and binning resulted in the reconstruction of 82 bacterial genomes from different redox regimes of estuary sediments. These genomes belong to 23 bacterial groups, including uncultured candidate phyla (for example, KSB1, TA06, and KD3-62) and three newly described phyla (White Oak River (WOR)-1, WOR-2, and WOR-3). The uncultured phyla are generally most abundant in the sulfate-methane transition (SMTZ) and methane-rich zones, and genomic data predict that they mediate essential biogeochemical processes of the estuarine environment, including organic carbon degradation and fermentation. Among the most abundant organisms in the sulfate-rich layer are novel Gammaproteobacteria that have genes for the oxidation of sulfur and the reduction of nitrate and nitrite. Interestingly, the terminal steps of denitrification (NO3 to N2O and then N2O to N2) are present in distinct bacterial populations.
Conclusions: This dataset extends our knowledge of the metabolic potential of several uncultured phyla. Within the sediments, there is redundancy in the genomic potential in different lineages, often distinct phyla, for essential biogeochemical processes. We were able to chart the flow of carbon and nutrients through the multiple geochemical layers of bacterial processing and reveal potential ecological interactions within the communities.
Keywords: Anaerobic respiration; Candidate phyla; Carbon; Estuary; Metagenome; Nitrogen; Sediment; Sulfate reduction; Sulfur.
Figures

References
-
- Whitaker RH, Liken GE. The biosphere and man. In: Leith H, Whitaker RH, editors. Primary productivity of the biosphere. N.Y: Spring-Verlag; 1975. pp. 305–28.
-
- Baker BJ, Dick GJ. Omic approaches in microbial ecology, charting the unknown. Microbe. 2013;8:353–9.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

