Selective inhibition of esophageal cancer cells by combination of HDAC inhibitors and Azacytidine
- PMID: 25923331
- PMCID: PMC4623041
- DOI: 10.1080/15592294.2015.1039216
Selective inhibition of esophageal cancer cells by combination of HDAC inhibitors and Azacytidine
Abstract
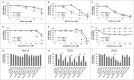
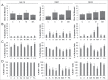
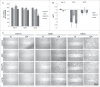
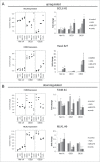
Esophageal cancers are highly aggressive tumors with poor prognosis despite some recent advances in surgical and radiochemotherapy treatment options. This study addressed the feasibility of drugs targeting epigenetic modifiers in esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) cells. We tested inhibition of histone deacetylases (HDACs) by SAHA, MS-275, and FK228, inhibition of DNA methyltransferases by Azacytidine (AZA) and Decitabine (DAC), and the effect of combination treatment using both types of drugs. The drug targets, HDAC1/2/3 and DNMT1, were expressed in normal esophageal epithelium and tumor cells of ESCC or EAC tissue specimens, as well as in non-neoplastic esophageal epithelial (Het-1A), ESCC (OE21, Kyse-270, Kyse-410), and EAC (OE33, SK-GT-4) cell lines. In vitro, HDAC activity, histone acetylation, and p21 expression were similarly affected in non-neoplastic, ESCC, and EAC cell lines post inhibitor treatment. Combined MS-275/AZA treatment, however, selectively targeted esophageal cancer cell lines by inducing DNA damage, cell viability loss, and apoptosis, and by decreasing cell migration. Non-neoplastic Het-1A cells were protected against HDACi (MS-275)/AZA treatment. RNA transcriptome analyses post MS-275 and/or AZA treatment identified novel regulated candidate genes (up: BCL6, Hes2; down: FAIM, MLKL), which were specifically associated with the treatment responses of esophageal cancer cells. In summary, combined HDACi/AZA treatment is efficient and selective for the targeting of esophageal cancer cells, despite similar target expression of normal and esophageal cancer epithelium, in vitro and in human esophageal carcinomas. The precise mechanisms of action of treatment responses involve novel candidate genes regulated by HDACi/AZA in esophageal cancer cells. Together, targeting of epigenetic modifiers in esophageal cancers may represent a potential future therapeutic approach.
Keywords: 5mC, 5-methylcytidine; AZA, Azacytidine; DAC, Decitabine; DNMT, DNA (cytosine-5)-methyltransferase; EAC, esophageal adenocarcinoma; ESCC, esophageal squamous cell carcinoma; FAIM, Fas apoptotic inhibitory molecule; GEJ, gastro-esophageal junction; H3Ac, histone H3 acetylation; H3K4me3, histone H3 trimethylation at lysine 4; H3K9Ac, histone 3 lysine 9 acetylation; HDAC, histone deacetylases; HDACi, HDAC inhibitor; Hes-2, Hairy and enhancer of split 2; SAHA, suberoylanilide hydroxamic acid; TSA, Trichostatin A; azacytidine/gene pathway regulation; epigenetics/HDAC inhibitor; esophageal cancer.
Figures




References
-
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003; 349:2241-52; PMID:14657432; http://dx.doi.org/ 10.1056/NEJMra035010 - DOI - PubMed
-
- Wijnhoven BP, Tran KT, Esterman A, Watson DI, Tilanus HW. An evaluation of prognostic factors and tumor staging of resected carcinoma of the esophagus. Ann Surg 2007; 245:717-25; PMID:17457164; http://dx.doi.org/ 10.1097/01.sla.0000251703.35919.02 - DOI - PMC - PubMed
-
- Cools-Lartigue J, Spicer J, Ferri LE. Current Status of Management of Malignant Disease: Current Management of Esophageal Cancer. J Gastrointest Surg 2015; PMID:25650163 - PubMed
-
- Makowiec F, Baier P, Kulemann B, Marjanovic G, Bronsert P, Zirlik K, Henke M, Hopt UT, Hoeppner J. Improved long-term survival after esophagectomy for esophageal cancer: influence of epidemiologic shift and neoadjuvant therapy. J Gastrointest Surg 2013; 17:1193-201; PMID:23636882; http://dx.doi.org/ 10.1007/s11605-013-2212-7 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
