Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma
- PMID: 25924914
- PMCID: PMC4529346
- DOI: 10.1002/cam4.451
Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma
Abstract
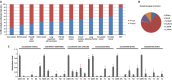
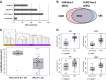
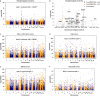
Altered patterns of DNA methylation are key features of cancer. Nasopharyngeal carcinoma (NPC) has the highest incidence in Southern China. Aberrant methylation at the promoter region of tumor suppressors is frequently reported in NPC; however, genome-wide methylation changes have not been comprehensively investigated. Therefore, we systematically analyzed methylome data in 25 primary NPC tumors and nontumor counterparts using a high-throughput approach with the Illumina HumanMethylation450 BeadChip. Comparatively, we examined the methylome data of 11 types of solid tumors collected by The Cancer Genome Atlas (TCGA). In NPC, the hypermethylation pattern was more dominant than hypomethylation and the majority of de novo methylated loci were within or close to CpG islands in tumors. The comparative methylome analysis reveals hypermethylation at chromosome 6p21.3 frequently occurred in NPC (false discovery rate; FDR=1.33 × 10(-9) ), but was less obvious in other types of solid tumors except for prostate and Epstein-Barr virus (EBV)-positive gastric cancer (FDR<10(-3) ). Bisulfite pyrosequencing results further confirmed the aberrant methylation at 6p in an additional patient cohort. Evident enrichment of the repressive mark H3K27me3 and active mark H3K4me3 derived from human embryonic stem cells were found at these regions, indicating both DNA methylation and histone modification function together, leading to epigenetic deregulation in NPC. Our study highlights the importance of epigenetic deregulation in NPC. Polycomb Complex 2 (PRC2), responsible for H3K27 trimethylation, is a promising therapeutic target. A key genomic region on 6p with aberrant methylation was identified. This region contains several important genes having potential use as biomarkers for NPC detection.
Keywords: Chromosome 6p; EBV; Illumina HumanMethylation450; methylome; nasopharyngeal carcinoma.
© 2015 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures


References
-
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. - PubMed
-
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

