Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA
- PMID: 25925567
- PMCID: PMC4446448
- DOI: 10.1093/nar/gkv415
Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA
Abstract
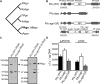
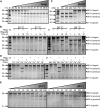
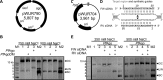
Functions of prokaryotic Argonautes (pAgo) have long remained elusive. Recently, Argonautes of the bacteria Rhodobacter sphaeroides and Thermus thermophilus were demonstrated to be involved in host defense. The Argonaute of the archaeon Pyrococcus furiosus (PfAgo) belongs to a different branch in the phylogenetic tree, which is most closely related to that of RNA interference-mediating eukaryotic Argonautes. Here we describe a functional and mechanistic characterization of PfAgo. Like the bacterial counterparts, archaeal PfAgo contributes to host defense by interfering with the uptake of plasmid DNA. PfAgo utilizes small 5'-phosphorylated DNA guides to cleave both single stranded and double stranded DNA targets, and does not utilize RNA as guide or target. Thus, with respect to function and specificity, the archaeal PfAgo resembles bacterial Argonautes much more than eukaryotic Argonautes. These findings demonstrate that the role of Argonautes is conserved through the bacterial and archaeal domains of life and suggests that eukaryotic Argonautes are derived from DNA-guided DNA-interfering host defense systems.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

