A novel sigma factor reveals a unique regulon controlling cell-specific recombination in Mycoplasma genitalium
- PMID: 25925568
- PMCID: PMC4446450
- DOI: 10.1093/nar/gkv422
A novel sigma factor reveals a unique regulon controlling cell-specific recombination in Mycoplasma genitalium
Abstract
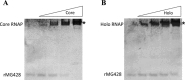
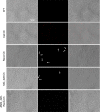
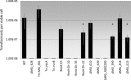
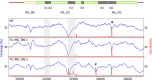
The Mycoplasma genitalium MG428 protein shows homology to members of the sigma-70 family of sigma factors. Herein, we found that MG428 activates transcription of recA, ruvA and ruvB as well as several genes with unknown function. Deletion of MG_428 or some of the up-regulated unknown genes led to severe recombination defects. Single cell analyses revealed that activation of the MG428-regulon is a rare event under laboratory growth conditions. A conserved sequence with sigma-70 promoter architecture (TTGTCA-N(18/19)-ATTWAT) was identified in the upstream region of all of the MG428-regulated genes or operons. Primer extension analyses demonstrated that transcription initiates immediately downstream of this sigma70-type promoter in a MG428-dependent manner. Furthermore, mutagenesis of the conserved -10 and -35 elements corroborated the requirement of these regions for promoter function. Therefore, a new mycoplasma promoter directs transcription of a unique recombination regulon. Additionally, MG428 was found to interact with the RNAP core enzyme, reinforcing the predicted role of this protein as an alternative sigma factor. Finally, our results indicate that MG428 contributes to the generation of genetic diversity in this model organism. Since recombination is an important mechanism to generate antigenic variation, MG428 emerges as a novel factor contributing to M. genitalium virulence.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M., et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. - PubMed
-
- Gibson D.G., Benders G.A., Andrews-Pfannkoch C., Denisova E.A., Baden-Tillson H., Zaveri J., Stockwell T.B., Brownley A., Thomas D.W., Algire M.A., et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

