NGL Viewer: a web application for molecular visualization
- PMID: 25925569
- PMCID: PMC4489237
- DOI: 10.1093/nar/gkv402
NGL Viewer: a web application for molecular visualization
Abstract
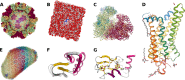
The NGL Viewer (http://proteinformatics.charite.de/ngl) is a web application for the visualization of macromolecular structures. By fully adopting capabilities of modern web browsers, such as WebGL, for molecular graphics, the viewer can interactively display large molecular complexes and is also unaffected by the retirement of third-party plug-ins like Flash and Java Applets. Generally, the web application offers comprehensive molecular visualization through a graphical user interface so that life scientists can easily access and profit from available structural data. It supports common structural file-formats (e.g. PDB, mmCIF) and a variety of molecular representations (e.g. 'cartoon, spacefill, licorice'). Moreover, the viewer can be embedded in other web sites to provide specialized visualizations of entries in structural databases or results of structure-related calculations.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

