The ribonuclease DIS3 promotes let-7 miRNA maturation by degrading the pluripotency factor LIN28B mRNA
- PMID: 25925570
- PMCID: PMC4446438
- DOI: 10.1093/nar/gkv387
The ribonuclease DIS3 promotes let-7 miRNA maturation by degrading the pluripotency factor LIN28B mRNA
Abstract
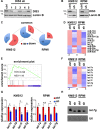
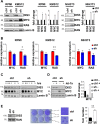
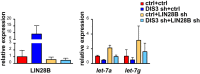
Multiple myeloma, the second most frequent hematologic tumor after lymphomas, is an incurable cancer. Recent sequencing efforts have identified the ribonuclease DIS3 as one of the most frequently mutated genes in this disease. DIS3 represents the catalytic subunit of the exosome, a macromolecular complex central to the processing, maturation and surveillance of various RNAs. miRNAs are an evolutionarily conserved class of small noncoding RNAs, regulating gene expression at post-transcriptional level. Ribonucleases, including Drosha, Dicer and XRN2, are involved in the processing and stability of miRNAs. However, the role of DIS3 on the regulation of miRNAs remains largely unknown. Here we found that DIS3 regulates the levels of the tumor suppressor let-7 miRNAs without affecting other miRNA families. DIS3 facilitates the maturation of let-7 miRNAs by reducing in the cytoplasm the RNA stability of the pluripotency factor LIN28B, a inhibitor of let-7 processing. DIS3 inactivation, through the increase of LIN28B and the reduction of mature let-7, enhances the translation of let-7 targets such as MYC and RAS leading to enhanced tumorigenesis. Our study establishes that the ribonuclease DIS3, targeting LIN28B, sustains the maturation of let-7 miRNAs and suggests the increased translation of critical oncogenes as one of the biological outcomes of DIS3 inactivation.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997;91:457–466. - PubMed
-
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. - PubMed
-
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

