Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development
- PMID: 25925669
- PMCID: PMC4515120
- DOI: 10.15252/embr.201439700
Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development
Abstract
Epigenetic modifications, such as DNA methylation and histone modifications, are dynamically altered predominantly in paternal pronuclei soon after fertilization. To identify which histone modifications are required for early embryonic development, we utilized histone K-M mutants, which prevent endogenous histone methylation at the mutated site. We prepared four single K-M mutants for histone H3.3, K4M, K9M, K27M, and K36M, and demonstrate that overexpression of H3.3 K4M in embryos before fertilization results in developmental arrest, whereas overexpression after fertilization does not affect the development. Furthermore, loss of H3K4 methylation decreases the level of minor zygotic gene activation (ZGA) predominantly in the paternal pronucleus, and we obtained similar results from knockdown of the H3K4 methyltransferase Mll3/4. We therefore conclude that H3K4 methylation, likely established by Mll3/4 at the early pronuclear stage, is essential for the onset of minor ZGA in the paternal pronucleus, which is necessary for subsequent preimplantation development in mice.
Keywords: K‐M mutant; Mll3/4; histone methylation; minor ZGA.
© 2015 The Authors.
Figures
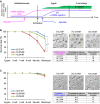
A Scheme of the experimental procedure. Magenta and blue lines indicate the estimated protein levels from injected mRNA. Green bars indicate H3K4me1/3 modification detected in paternal genome.
B, C Developmental rate of zygotes overexpressing H3.3 WT, H3.3 K4M, and H3.3 K4R before (B) or after (C) fertilization. Each figure shows survival curves (left), representative pictures (right top), and developmental rates (right bottom). Survival curves are presented as average percentages ± s.d. from three independent experiments. Chi-square test was performed for analyses of embryos through development to the blastocyst stage. Scale bars, 50 μm. *P < 0.01.
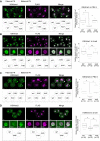
A–D Immunostaining against H3K4me1 and H3K4me3 was performed for PN zygotes (A, C) and 2-cell embryos (B, D). Left top pictures are representative images, and left bottom schemes indicate which type of H3.3 mRNA was injected into embryos corresponding to the representative images. Left middle pictures in (B) and (D) are magnified images of the nucleus in each 2-cell embryo. Right graphs are boxplots for relative intensities of H3K4me1 or H3K4me3 from three independent experiments. Statistical analyses performed were the Games–Howell test (A), Tukey–Kramer test (B, D), and Steel–Dwass test (C). Scale bars, 20 μm. *P < 0.01; **P < 0.05.
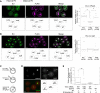
A, B Left pictures are representative fluorescent images of zygotes overexpressing H3.3 WT, H3.3 K4M, and H3.3 K4R after EU treatment for PN zygotes (A) and 2-cell embryos (B). Left bottom schemes indicate which type of H3.3 mRNA was injected into embryos corresponding to the representative images. Right graphs are boxplots for relative intensities of EU from four independent experiments.
C Scheme of the experimental procedure for (D) and (E).
D Representative image of PN4–5 zygotes expressing sperm-derived H4-Venus. Dotted circles indicate pronuclei according to the DAPI staining (red pseudo-color). PB: polar bodies.
E Relative expression levels of H4-Venus by single-cell RT–qPCR. Sample numbers are as indicated; the normalization method is described in the Supplementary Materials and Methods.
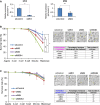
A mRNA expression of Mll3 and Mll4 was examined for 2-cell embryos which were microinjected with siMll3 and siMll4, respectively. Data are presented as means ± s.d. from three independent experiments. The y-axis indicates relative expression levels with respect to the value of siControl-injected embryo, set to 1 for each group. *P < 0.01; **P < 0.05
B, C Developmental rate of zygotes microinjected with siControl, siMll3, siMll3&4, and siMll4 before (B) or after (C) fertilization. Each figure shows survival curves (left), representative pictures (right top), and developmental rates (right bottom). Survival curves are presented as average percentages ± s.d. from three independent experiments. Chi-square test was performed for analyses of embryos that developed to the blastocyst stage. Scale bars, 50 μm. *P < 0.01; **P < 0.05.

A, B Immunostaining against H3K4me1 and H3K27ac (A) and EU treatment (B) were performed for PN zygotes. In each figure, top pictures are representative images, and bottom schemes indicate which type of siRNA was injected into embryos corresponding to the representative images.
C–E Boxplots for relative intensities of H3K4me1 and H3K27ac or EU from three independent experiments for each expression as in (A) and (B). Statistical analyses performed were the Steel–Dwass test (H3K4me1) and Tukey–Kramer test (H3K27ac and EU). Scale bars, 20 μm. *P < 0.01.
Comment in
-
KM mutant highlights enhancers in minor ZGA.Cell Cycle. 2015;14(16):2541-2. doi: 10.1080/15384101.2015.1060774. Epub 2015 Jun 11. Cell Cycle. 2015. PMID: 26065875 Free PMC article. No abstract available.
References
-
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. - PubMed
-
- Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46:317–320. - PubMed
-
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. - PubMed
-
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

