The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae
- PMID: 25926236
- PMCID: PMC4405625
- DOI: 10.1128/CMR.00116-14
The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae
Abstract
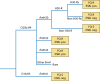
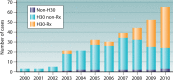
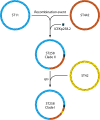
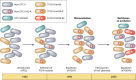
Escherichia coli sequence type 131 (ST131) and Klebsiella pneumoniae ST258 emerged in the 2000s as important human pathogens, have spread extensively throughout the world, and are responsible for the rapid increase in antimicrobial resistance among E. coli and K. pneumoniae strains, respectively. E. coli ST131 causes extraintestinal infections and is often fluoroquinolone resistant and associated with extended-spectrum β-lactamase production, especially CTX-M-15. K. pneumoniae ST258 causes urinary and respiratory tract infections and is associated with carbapenemases, most often KPC-2 and KPC-3. The most prevalent lineage within ST131 is named fimH30 because it contains the H30 variant of the type 1 fimbrial adhesin gene, and recent molecular studies have demonstrated that this lineage emerged in the early 2000s and was then followed by the rapid expansion of its sublineages H30-R and H30-Rx. K. pneumoniae ST258 comprises 2 distinct lineages, namely clade I and clade II. Moreover, it seems that ST258 is a hybrid clone that was created by a large recombination event between ST11 and ST442. Epidemic plasmids with blaCTX-M and blaKPC belonging to incompatibility group F have contributed significantly to the success of these clones. E. coli ST131 and K. pneumoniae ST258 are the quintessential examples of international multidrug-resistant high-risk clones.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland.
-
- Reuter S, Ellington MJ, Cartwright EJ, Koser CU, Torok ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

