Polyester: simulating RNA-seq datasets with differential transcript expression
- PMID: 25926345
- PMCID: PMC4635655
- DOI: 10.1093/bioinformatics/btv272
Polyester: simulating RNA-seq datasets with differential transcript expression
Abstract
Motivation: Statistical methods development for differential expression analysis of RNA sequencing (RNA-seq) requires software tools to assess accuracy and error rate control. Since true differential expression status is often unknown in experimental datasets, artificially constructed datasets must be utilized, either by generating costly spike-in experiments or by simulating RNA-seq data.
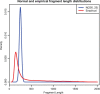
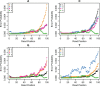
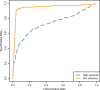
Results: Polyester is an R package designed to simulate RNA-seq data, beginning with an experimental design and ending with collections of RNA-seq reads. Its main advantage is the ability to simulate reads indicating isoform-level differential expression across biological replicates for a variety of experimental designs. Data generated by Polyester is a reasonable approximation to real RNA-seq data and standard differential expression workflows can recover differential expression set in the simulation by the user.
Availability and implementation: Polyester is freely available from Bioconductor (http://bioconductor.org/).
Contact: jtleek@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
SimSeq: a nonparametric approach to simulation of RNA-sequence datasets.Bioinformatics. 2015 Jul 1;31(13):2131-40. doi: 10.1093/bioinformatics/btv124. Epub 2015 Feb 26. Bioinformatics. 2015. PMID: 25725090 Free PMC article.
-
rSeqNP: a non-parametric approach for detecting differential expression and splicing from RNA-Seq data.Bioinformatics. 2015 Jul 1;31(13):2222-4. doi: 10.1093/bioinformatics/btv119. Epub 2015 Feb 24. Bioinformatics. 2015. PMID: 25717189 Free PMC article.
-
SPARTA: Simple Program for Automated reference-based bacterial RNA-seq Transcriptome Analysis.BMC Bioinformatics. 2016 Feb 4;17:66. doi: 10.1186/s12859-016-0923-y. BMC Bioinformatics. 2016. PMID: 26847232 Free PMC article.
-
Differential Expression Analysis of RNA-seq Reads: Overview, Taxonomy, and Tools.IEEE/ACM Trans Comput Biol Bioinform. 2020 Mar-Apr;17(2):566-586. doi: 10.1109/TCBB.2018.2873010. Epub 2018 Oct 1. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30281477 Review.
-
From RNA-seq reads to differential expression results.Genome Biol. 2010;11(12):220. doi: 10.1186/gb-2010-11-12-220. Epub 2010 Dec 22. Genome Biol. 2010. PMID: 21176179 Free PMC article. Review.
Cited by
-
The Bellerophon pipeline, improving de novo transcriptomes and removing chimeras.Ecol Evol. 2019 Aug 17;9(18):10513-10521. doi: 10.1002/ece3.5571. eCollection 2019 Sep. Ecol Evol. 2019. PMID: 31624564 Free PMC article.
-
The value of genotype-specific reference for transcriptome analyses in barley.Life Sci Alliance. 2022 Apr 22;5(8):e202101255. doi: 10.26508/lsa.202101255. Print 2022 Aug. Life Sci Alliance. 2022. PMID: 35459738 Free PMC article.
-
Combining Multiple RNA-Seq Data Analysis Algorithms Using Machine Learning Improves Differential Isoform Expression Analysis.Methods Protoc. 2021 Sep 27;4(4):68. doi: 10.3390/mps4040068. Methods Protoc. 2021. PMID: 34698224 Free PMC article.
-
Spatially Resolved Expression of Transposable Elements in Disease and Somatic Tissue with SpatialTE.Int J Mol Sci. 2021 Dec 20;22(24):13623. doi: 10.3390/ijms222413623. Int J Mol Sci. 2021. PMID: 34948421 Free PMC article.
-
SeqNet: An R Package for Generating Gene-Gene Networks and Simulating RNA-Seq Data.J Stat Softw. 2021 Jul;98(12):10.18637/jss.v098.i12. doi: 10.18637/jss.v098.i12. Epub 2021 Jul 10. J Stat Softw. 2021. PMID: 34321962 Free PMC article.
References
-
- AC’t Hoen P., et al. (2013). Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat. Biotechnol , 31, 1015–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

