Identification of a Novel Hepacivirus in Domestic Cattle from Germany
- PMID: 25926652
- PMCID: PMC4473572
- DOI: 10.1128/JVI.00534-15
Identification of a Novel Hepacivirus in Domestic Cattle from Germany
Abstract
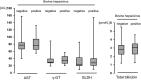
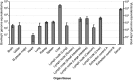
Hepatitis C virus (HCV) continues to represent one of the most significant threats to human health. In recent years, HCV-related sequences have been found in bats, rodents, horses, and dogs, indicating a widespread distribution of hepaciviruses among animals. By applying unbiased high-throughput sequencing, a novel virus of the genus Hepacivirus was discovered in a bovine serum sample. De novo assembly yielded a nearly full-length genome coding for a polyprotein of 2,779 amino acids. Phylogenetic analysis confirmed that the virus represents a novel species within the genus Hepacivirus. Viral RNA screening determined that 1.6% (n = 5) of 320 individual animals and 3.2% (n = 5) of 158 investigated cattle herds in Germany were positive for bovine hepacivirus. Repeated reverse transcription-PCR (RT-PCR) analyses of animals from one dairy herd proved that a substantial percentage of cows were infected, with some of them being viremic for over 6 months. Clinical and postmortem examination revealed no signs of disease, including liver damage. Interestingly, quantitative RT-PCR from different organs and tissues, together with the presence of an miR-122 binding site in the viral genome, strongly suggests a liver tropism for bovine hepacivirus, making this novel virus a promising animal model for HCV infections in humans.
Importance: Livestock animals act as important sources for emerging pathogens. In particular, their large herd size and the existence of multiple ways of direct and food-borne infection routes emphasize their role as virus reservoirs. Apart from the search for novel viruses, detailed characterization of these pathogens is indispensable in the context of risk analysis. Here, we describe the identification of a novel HCV-like virus in cattle. In addition, determination of the prevalence and of the course of infection in cattle herds provides valuable insights into the biology of this novel virus. The results presented here form a basis for future studies targeting viral pathogenesis of bovine hepaciviruses and their potential to establish zoonotic infections.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

