An epitope tag alters phosphoglycerate dehydrogenase structure and impairs ability to support cell proliferation
- PMID: 25926973
- PMCID: PMC4414297
- DOI: 10.1186/s40170-015-0131-7
An epitope tag alters phosphoglycerate dehydrogenase structure and impairs ability to support cell proliferation
Abstract
Background: The gene encoding the serine biosynthesis pathway enzyme PHGDH is located in a region of focal genomic copy number gain in human cancers. Cells with PHGDH amplification are dependent on enzyme expression for proliferation. However, dependence on increased PHGDH expression extends beyond production of serine alone, and further studies of PHGDH function are necessary to elucidate its role in cancer cells. These studies will require a physiologically relevant form of the enzyme for experiments using engineered cell lines and recombinant protein.
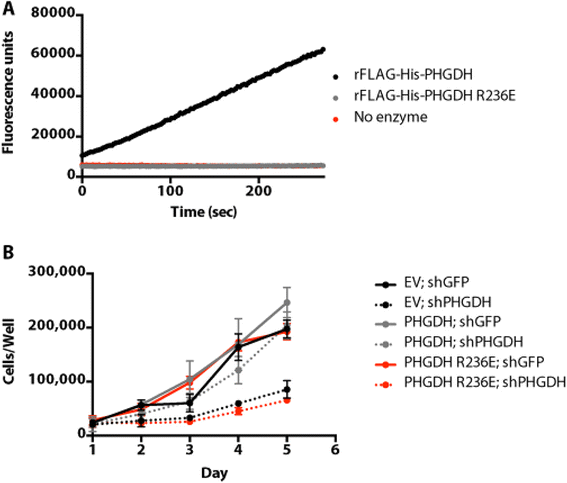
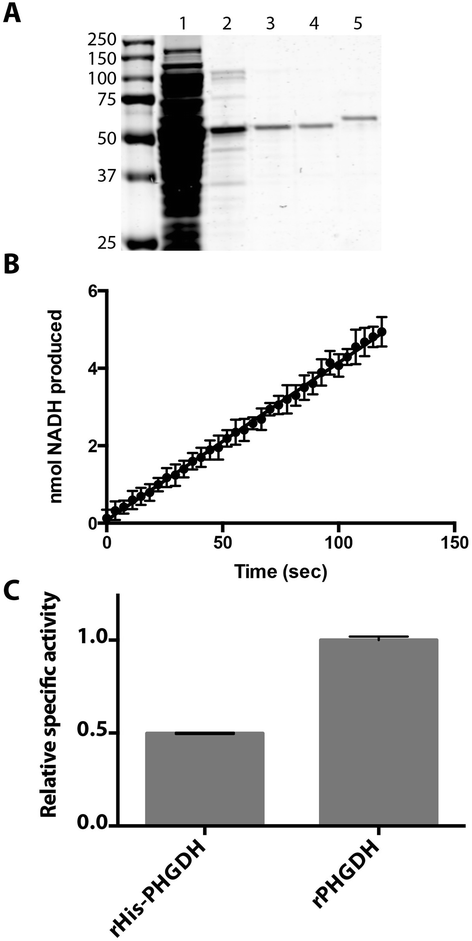
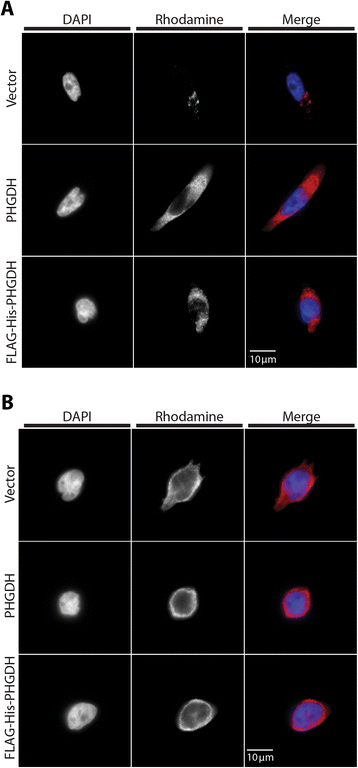
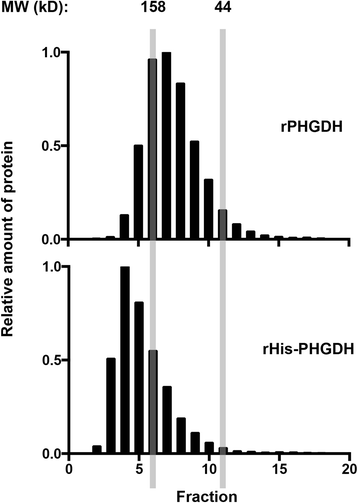
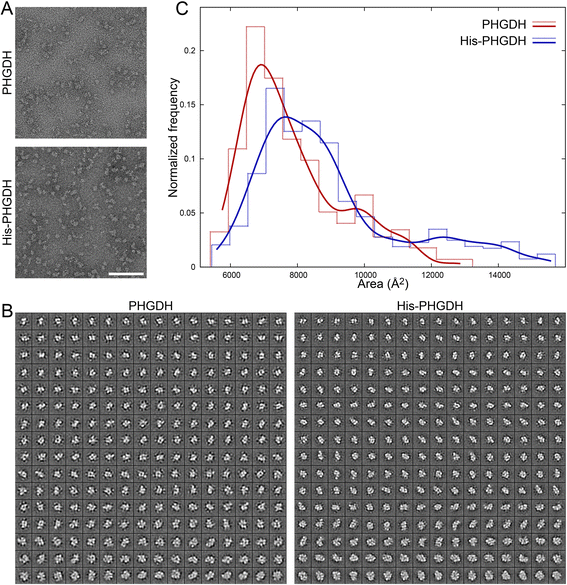
Results: The addition of an N-terminal epitope tag to PHGDH abolished the ability to support proliferation of PHGDH-amplified cells despite retention of some activity to convert 3-PG to PHP. Introducing an R236E mutation into PHGDH eliminates enzyme activity, and this catalytically inactive enzyme cannot support proliferation of PHGDH-dependent cells, arguing that canonical enzyme activity is required. Tagged and untagged PHGDH exhibit the same intracellular localization and ability to produce D-2-hydroxyglutarate (D-2HG), an error product of PHGDH, arguing that neither mislocalization nor loss of D-2HG production explains the inability of epitope-tagged PHGDH to support proliferation. To enable studies of PHGDH function, we report a method to purify recombinant PHGDH and found that untagged enzyme activity was greater than N-terminally tagged enzyme. Analysis of tagged and untagged PHGDH using size exclusion chromatography and electron microscopy found that an N-terminal epitope tag alters enzyme structure.
Conclusions: Purification of untagged recombinant PHGDH eliminates the need to use an epitope tag for enzyme studies. Furthermore, while tagged PHGDH retains some ability to convert 3PG to PHP, the structural alterations caused by including an epitope tag disrupts the ability of PHGDH to sustain cancer cell proliferation.
Keywords: Cancer Metabolism; Epitope tag; Phosphoglycerate dehydrogenase; Serine metabolism; Serine synthesis.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

