PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD)
- PMID: 25927346
- PMCID: PMC4615115
- DOI: 10.1080/15384101.2015.1033595
PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD)
Abstract
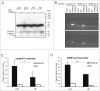
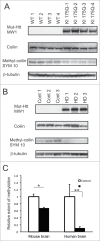
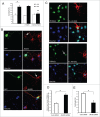
Abnormal protein interactions of mutant huntingtin (Htt) triggered by polyglutamine expansion are thought to mediate Huntington's disease (HD) pathogenesis. Here, we explored a functional interaction of Htt with protein arginine methyltransferase 5 (PRMT5), an enzyme mediating symmetrical dimethylation of arginine (sDMA) of key cellular proteins, including histones, and spliceosomal Sm proteins. Gene transcription and RNA splicing are impaired in HD. We demonstrated PRMT5 and Htt interaction and their co-localization in transfected neurons and in HD brain. As a result of this interaction, normal (but to a lesser extend mutant) Htt stimulated PRMT5 activity in vitro. SDMA of histones H2A and H4 was reduced in the presence of mutant Htt in primary cultured neurons and in HD brain, consistent with a demonstrated reduction in R3Me2s occupancy at the transcriptionally repressed promoters in HD brain. SDMA of another PRMT5 substrate, Cajal body marker coilin, was also reduced in the HD mouse model and in human HD brain. Finally, compensation of PRMT5 deficiency by ectopic expression of PRMT5/MEP50 complexes, or by the knock-down of H4R3Me2 demethylase JMJD6, reversed the toxic effects of mutant Htt in primary cortical neurons, suggesting that PRMT5 deficiency may mediate, at least in part, HD pathogenesis. These studies revealed a potential new mechanism for disruption of gene expression and RNA processing in HD, involving a loss of normal function of Htt in facilitation of PRMT5, supporting the idea that epigenetic regulation of gene transcription may be involved in HD and highlighting symmetric dimethylation of arginine as potential new therapeutic target.
Keywords: BDNF, brain-derived neurotrophic factor; CB, Cajal body; ChIP, the chromatin immunoprecipitation; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; HD, Huntington's disease; HEK, human embryonic kidney; Htt, huntingtin; Huntington's disease mechanism; IP, immunoprecipitation; IgG, immunoglobulin; PIC, protease inhibitors cocktail; PRMT5, protein arginine methyltransferase; RNA processing; SMN, survival of motor neurons; Sm proteins, spleceosomal small nuclear ribonucleoproteins; gene transcription; huntingtin; neurodegeneration; polyQ, polyglutamine; protein interactions; protein methylation; sDMA, symmetrical arginine dimethylation; snRNPs, small nuclear ribonucleoprotein particles.
Figures

Similar articles
-
Huntingtin-mediated axonal transport requires arginine methylation by PRMT6.Cell Rep. 2021 Apr 13;35(2):108980. doi: 10.1016/j.celrep.2021.108980. Cell Rep. 2021. PMID: 33852844 Free PMC article.
-
Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington's disease.Cell Death Differ. 2004 Apr;11(4):424-38. doi: 10.1038/sj.cdd.4401358. Cell Death Differ. 2004. PMID: 14713958
-
A series of N-terminal epitope tagged Hdh knock-in alleles expressing normal and mutant huntingtin: their application to understanding the effect of increasing the length of normal Huntingtin's polyglutamine stretch on CAG140 mouse model pathogenesis.Mol Brain. 2012 Aug 14;5:28. doi: 10.1186/1756-6606-5-28. Mol Brain. 2012. PMID: 22892315 Free PMC article.
-
Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease.Biochem Soc Trans. 2009 Dec;37(Pt 6):1270-5. doi: 10.1042/BST0371270. Biochem Soc Trans. 2009. PMID: 19909260 Review.
-
Wild-type huntingtin plays a role in brain development and neuronal survival.Mol Neurobiol. 2003 Dec;28(3):259-76. doi: 10.1385/MN:28:3:259. Mol Neurobiol. 2003. PMID: 14709789 Review.
Cited by
-
The Structure and Functions of PRMT5 in Human Diseases.Life (Basel). 2021 Oct 12;11(10):1074. doi: 10.3390/life11101074. Life (Basel). 2021. PMID: 34685445 Free PMC article. Review.
-
The Methylation Status of the Epigenome: Its Emerging Role in the Regulation of Tumor Angiogenesis and Tumor Growth, and Potential for Drug Targeting.Cancers (Basel). 2018 Aug 10;10(8):268. doi: 10.3390/cancers10080268. Cancers (Basel). 2018. PMID: 30103412 Free PMC article. Review.
-
Correction of Huntington's Disease Phenotype by Genistein-Induced Autophagy in the Cellular Model.Neuromolecular Med. 2018 Mar;20(1):112-123. doi: 10.1007/s12017-018-8482-1. Epub 2018 Feb 12. Neuromolecular Med. 2018. PMID: 29435951 Free PMC article.
-
Protein Arginine Methyltransferases from Regulatory Function to Clinical Implication in Central Nervous System.Cell Mol Neurobiol. 2025 May 14;45(1):41. doi: 10.1007/s10571-025-01546-0. Cell Mol Neurobiol. 2025. PMID: 40366461 Free PMC article. Review.
-
Histone Methylation Regulation in Neurodegenerative Disorders.Int J Mol Sci. 2021 Apr 28;22(9):4654. doi: 10.3390/ijms22094654. Int J Mol Sci. 2021. PMID: 33925016 Free PMC article. Review.
References
-
- A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes . The Huntington's Disease Collaborative Research Group. Cell 1993; 72:971-83; PMID:8458085; http://dx.doi.org/10.1016/0092-8674(93)90585-E - DOI - PubMed
-
- Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS, et al.. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol 2014; 10:204-16; PMID:24614516; http://dx.doi.org/10.1038/nrneurol.2014.24 - DOI - PubMed
-
- Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol 2011; 10:83-98; PMID:21163446; http://dx.doi.org/10.1016/S1474-4422(10)70245-3 - DOI - PubMed
-
- Walker FO . Huntington's disease. Lancet 2007; 369:218-28; PMID:17240289; http://dx.doi.org/10.1016/S0140-6736(07)60111-1 - DOI - PubMed
-
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum Mol Genet 1999; 8:1647-55; PMID:10441327; http://dx.doi.org/10.1093/hmg/8.9.1647 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
