Clinical evaluation of commercial nucleic acid amplification tests in patients with suspected sepsis
- PMID: 25928122
- PMCID: PMC4419503
- DOI: 10.1186/s12879-015-0938-4
Clinical evaluation of commercial nucleic acid amplification tests in patients with suspected sepsis
Abstract
Background: Sepsis is a serious medical condition requiring timely administered, appropriate antibiotic therapy. Blood culture is regarded as the gold standard for aetiological diagnosis of sepsis, but it suffers from low sensitivity and long turnaround time. Thus, nucleic acid amplification tests (NAATs) have emerged to shorten the time to identification of causative microbes. The aim of the present study was to evaluate the clinical utility in everyday practice in the emergency department of two commercial NAATs in patients suspected with sepsis.
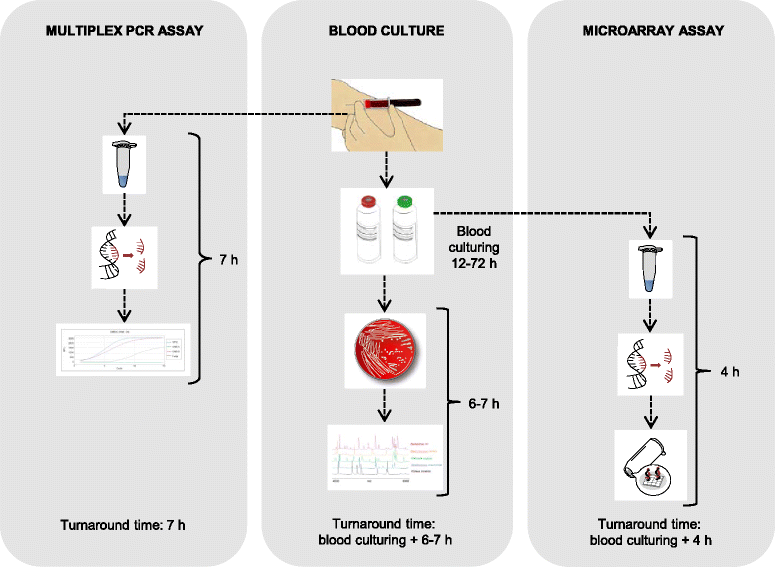
Methods: During a six-week period, blood samples were collected consecutively from all adult patients admitted to the general emergency department for suspicion of a community-onset sepsis and treated with intravenous antibiotics. Along with conventional blood cultures, multiplex PCR (Magicplex™) was performed on whole blood specimens whereas portions from blood culture bottles were used for analysis by microarray-based assay (Prove-it™). The aetiological significance of identified organisms was determined by two infectious disease physicians based on clinical presentation and expected pathogenicity.
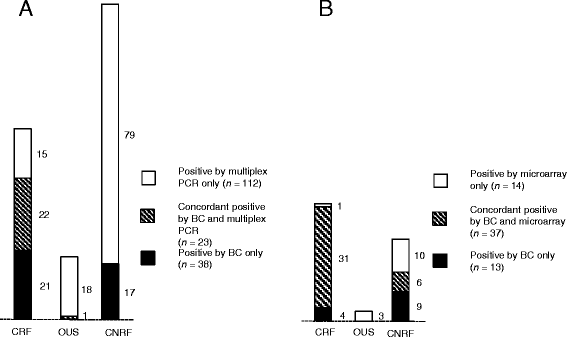
Results: Among 382 episodes of suspected sepsis, clinically relevant microbes were detected by blood culture in 42 episodes (11%), by multiplex PCR in 37 episodes (9.7%), and by microarray in 32 episodes (8.4%). Although moderate agreement with blood culture (kappa 0.50), the multiplex PCR added diagnostic value by timely detection of 15 clinically relevant findings in blood culture-negative specimens. Results of the microarray corresponded very well to those of blood culture (kappa 0.90), but were available just marginally prior to blood culture results.
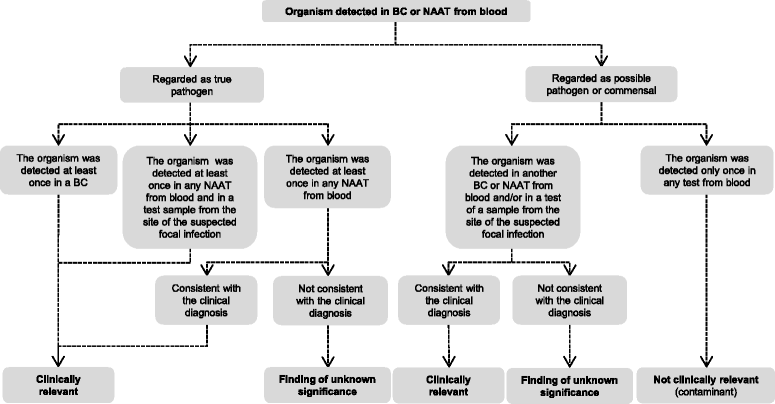
Conclusions: The use of NAATs on whole blood specimens in adjunct to current culture-based methods provides a clinical add-on value by allowing for detection of organisms missed by blood culture. However, the aetiological significance of findings detected by NAATs should be interpreted with caution as the high analytical sensitivity may add findings that do not necessarily corroborate with the clinical diagnosis.
Figures
Similar articles
-
Evaluation of the Punch-it™ NA-Sample kit for detecting microbial DNA in blood culture bottles using PCR-reverse blot hybridization assay.J Microbiol Methods. 2016 Sep;128:24-30. doi: 10.1016/j.mimet.2016.06.001. Epub 2016 Jun 2. J Microbiol Methods. 2016. PMID: 27263831
-
Performance of PCR-REBA assay for screening and identifying pathogens directly in whole blood of patients with suspected sepsis.J Appl Microbiol. 2015 Nov;119(5):1433-42. doi: 10.1111/jam.12941. Epub 2015 Oct 7. J Appl Microbiol. 2015. PMID: 26299262
-
Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit.Crit Care. 2010;14(4):R159. doi: 10.1186/cc9234. Epub 2010 Aug 24. Crit Care. 2010. PMID: 20731880 Free PMC article.
-
Sepsis Pathogen Identification.J Lab Autom. 2015 Oct;20(5):539-61. doi: 10.1177/2211068214567345. Epub 2015 Jan 28. J Lab Autom. 2015. PMID: 25631157 Review.
-
[Molecular biological sepsis diagnostic using multiplex PCR in surgical intensive care as suitable alternative to conventional microbial culture - a representative overview].Zentralbl Chir. 2011 Apr;136(2):135-42. doi: 10.1055/s-0031-1271407. Epub 2011 Apr 5. Zentralbl Chir. 2011. PMID: 21469038 Review. German.
Cited by
-
A retrospective evaluation of a multiplex polymerase chain reaction test directly applied to blood for the management of sepsis in the critically ill.South Afr J Crit Care. 2021 Dec 31;37(3):10.7196/SAJCC.2021.v37i3.495. doi: 10.7196/SAJCC.2021.v37i3.495. eCollection 2021. South Afr J Crit Care. 2021. PMID: 35517850 Free PMC article.
-
The Enterococcus: a Model of Adaptability to Its Environment.Clin Microbiol Rev. 2019 Jan 30;32(2):e00058-18. doi: 10.1128/CMR.00058-18. Print 2019 Mar 20. Clin Microbiol Rev. 2019. PMID: 30700430 Free PMC article. Review.
-
Genotypic Characterization of Clinical Klebsiella spp. Isolates Collected From Patients With Suspected Community-Onset Sepsis, Sweden.Front Microbiol. 2021 Apr 30;12:640408. doi: 10.3389/fmicb.2021.640408. eCollection 2021. Front Microbiol. 2021. PMID: 33995300 Free PMC article.
-
The Clinical Challenge of Sepsis Identification and Monitoring.PLoS Med. 2016 May 17;13(5):e1002022. doi: 10.1371/journal.pmed.1002022. eCollection 2016 May. PLoS Med. 2016. PMID: 27187803 Free PMC article.
-
Benchmarking of two bioinformatic workflows for the analysis of whole-genome sequenced Staphylococcus aureus collected from patients with suspected sepsis.BMC Infect Dis. 2023 Jan 20;23(1):39. doi: 10.1186/s12879-022-07977-0. BMC Infect Dis. 2023. PMID: 36670352 Free PMC article.
References
-
- Davies A, Green C, Hutton J. Severe sepsis: a European estimate of the burden of disease in IC. Intensive Care Med. 2001;27(Suppl 2):S284.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

