RNA-Seq Uncovers SNPs and Alternative Splicing Events in Asian Lotus (Nelumbo nucifera)
- PMID: 25928215
- PMCID: PMC4416007
- DOI: 10.1371/journal.pone.0125702
RNA-Seq Uncovers SNPs and Alternative Splicing Events in Asian Lotus (Nelumbo nucifera)
Abstract
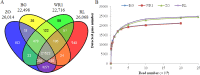
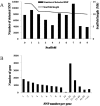
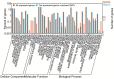
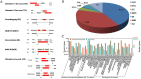
RNA-Seq is an efficient way to comprehensively identify single nucleotide polymorphisms (SNPs) and alternative splicing (AS) events from the expressed genes. In this study, we conducted transcriptome sequencing of four Asian lotus (Nelumbo nucifera) cultivars using Illumina HiSeq2000 platform to identify SNPs and AS events in lotus. A total of 505 million pair-end RNA-Seq reads were generated from four cultivars, of which 86% were mapped to the lotus reference genome. Using the four sets of data together, a total of 357,689 putative SNPs were identified with an average density of one SNP per 2.2 kb. These SNPs were located in 1,253 scaffolds and 15,016 expressed genes. A/G and C/T were the two major types of SNPs in the Asian lotus transcriptome. In parallel, a total of 177,540 AS events were detected in the four cultivars and were distributed in 64% of the expressed genes of lotus. The predominant type of AS events was alternative 5' first exon, which accounted for 41.2% of all the observed AS events, and exon skipping only accounted for 4.3% of all AS. Gene Ontology analysis was conducted to analyze the function of the genes containing SNPs and AS events. Validation of selected SNPs and AS events revealed that 74% of SNPs and 80% of AS events were reliable, which indicates that RNA-Seq is an efficient approach to uncover gene-associated SNPs and AS events. A large number of SNPs and AS events identified in our study will facilitate further genetic and functional genomics research in lotus.
Conflict of interest statement
Figures



Similar articles
-
The Latest Studies on Lotus (Nelumbo nucifera)-an Emerging Horticultural Model Plant.Int J Mol Sci. 2019 Jul 27;20(15):3680. doi: 10.3390/ijms20153680. Int J Mol Sci. 2019. PMID: 31357582 Free PMC article. Review.
-
Studies on Lotus Genomics and the Contribution to Its Breeding.Int J Mol Sci. 2022 Jun 30;23(13):7270. doi: 10.3390/ijms23137270. Int J Mol Sci. 2022. PMID: 35806274 Free PMC article. Review.
-
Construction of a high-density, high-quality genetic map of cultivated lotus (Nelumbo nucifera) using next-generation sequencing.BMC Genomics. 2016 Jun 17;17:466. doi: 10.1186/s12864-016-2781-4. BMC Genomics. 2016. PMID: 27317430 Free PMC article.
-
Genome-Wide Identification of SSR and SNP Markers Based on Whole-Genome Re-Sequencing of a Thailand Wild Sacred Lotus (Nelumbo nucifera).PLoS One. 2015 Nov 25;10(11):e0143765. doi: 10.1371/journal.pone.0143765. eCollection 2015. PLoS One. 2015. PMID: 26606530 Free PMC article.
-
Comparative population genomics reveals genetic divergence and selection in lotus, Nelumbo nucifera.BMC Genomics. 2020 Feb 11;21(1):146. doi: 10.1186/s12864-019-6376-8. BMC Genomics. 2020. PMID: 32046648 Free PMC article.
Cited by
-
The Latest Studies on Lotus (Nelumbo nucifera)-an Emerging Horticultural Model Plant.Int J Mol Sci. 2019 Jul 27;20(15):3680. doi: 10.3390/ijms20153680. Int J Mol Sci. 2019. PMID: 31357582 Free PMC article. Review.
-
Studies on Lotus Genomics and the Contribution to Its Breeding.Int J Mol Sci. 2022 Jun 30;23(13):7270. doi: 10.3390/ijms23137270. Int J Mol Sci. 2022. PMID: 35806274 Free PMC article. Review.
-
Comparative genomics and transcriptome analysis of Lactobacillus rhamnosus ATCC 11443 and the mutant strain SCT-10-10-60 with enhanced L-lactic acid production capacity.Mol Genet Genomics. 2018 Feb;293(1):265-276. doi: 10.1007/s00438-017-1379-0. Epub 2017 Nov 20. Mol Genet Genomics. 2018. PMID: 29159508
-
Gene-based SNP identification and validation in soybean using next-generation transcriptome sequencing.Mol Genet Genomics. 2018 Jun;293(3):623-633. doi: 10.1007/s00438-017-1410-5. Epub 2017 Dec 27. Mol Genet Genomics. 2018. PMID: 29280001
-
Metabolomic and Proteomic Profiles Reveal the Dynamics of Primary Metabolism during Seed Development of Lotus (Nelumbo nucifera).Front Plant Sci. 2016 Jun 7;7:750. doi: 10.3389/fpls.2016.00750. eCollection 2016. Front Plant Sci. 2016. PMID: 27375629 Free PMC article.
References
-
- Zhang XY, Chen LQ, Wang QC. New lotus flower cultivars in China Beijing: China Forestry Publishing House; 2011. pp. 278.
-
- Shen-Miller J. Sacred lotus, the long-living fruits of China Antique. Seed Science Research. 2002; 12: 131–143.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

