Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model
- PMID: 25928420
- PMCID: PMC4415773
- DOI: 10.1371/journal.pone.0124504
Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model
Abstract
Background and aims: It is known that postnatal functional maturation of the small intestine is facilitated by microbial colonization of the gut. Preterm infants exhibit defects in gut maturation, weak innate immunity against intestinal infection and increased susceptibility to inflammatory disorders, all of which may be related to the inappropriate microbial colonization of their immature intestines. The earliest microbes to colonize the preterm infant gut encounter a naïve, immature intestine. Thus this earliest microbiota potentially has the greatest opportunity to fundamentally influence intestinal development and immune function. The aim of this study was to characterize the effect of early microbial colonization on global gene expression in the distal small intestine during postnatal gut development.
Methods: Gnotobiotic mouse models with experimental colonization by early (prior to two weeks of life) intestinal microbiota from preterm human infants were utilized. Microarray analysis was used to assess global gene expression in the intestinal epithelium.
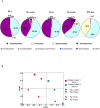
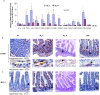
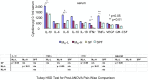
Results and conclusion: Multiple intestinal genes involved in metabolism, cell cycle regulation, cell-cell or cell-extracellular matrix communication, and immune function are developmental- and intestinal microbiota- regulated. Using a humanized gnotobiotic mouse model, we demonstrate that certain early preterm infant microbiota from prior to 2 weeks of life specifically induce increased NF-κB activation and a phenotype of increased inflammation whereas other preterm microbiota specifically induce decreased NF-κB activation. These fundamental differences correlate with altered clinical outcomes and suggest the existence of optimal early microbial communities to improve health outcomes.
Conflict of interest statement
Figures



References
-
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R (2004) Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell 118: 229–241. - PubMed
-
- Cario E, Gerken G, Podolsky DK (2007) Toll-Like Receptor 2 Controls Mucosal Inflammation by Regulating Epithelial Barrier Function. Gastroenterology 132: 1359–1374. - PubMed
-
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI (2001) Molecular Analysis of Commensal Host-Microbial Relationships in the Intestine. Science 291: 881–884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

