Elviz - exploration of metagenome assemblies with an interactive visualization tool
- PMID: 25928663
- PMCID: PMC4432942
- DOI: 10.1186/s12859-015-0566-4
Elviz - exploration of metagenome assemblies with an interactive visualization tool
Abstract
Background: Metagenomics, the sequencing of DNA collected from an entire microbial community, enables the study of natural microbial consortia in their native habitats. Metagenomics studies produce huge volumes of data, including both the sequences themselves and metadata describing their abundance, assembly, predicted functional characteristics and environmental parameters. The ability to explore these data visually is critically important to meaningful biological interpretation. Current genomics applications cannot effectively integrate sequence data, assembly metadata, and annotation to support both genome and community-level inquiry.
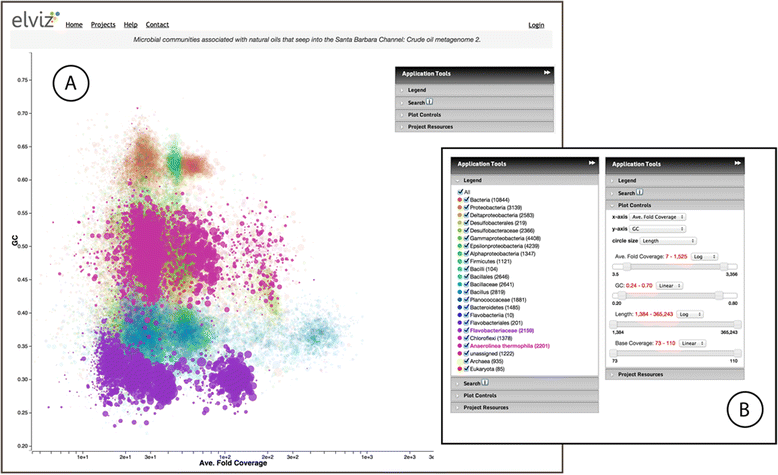
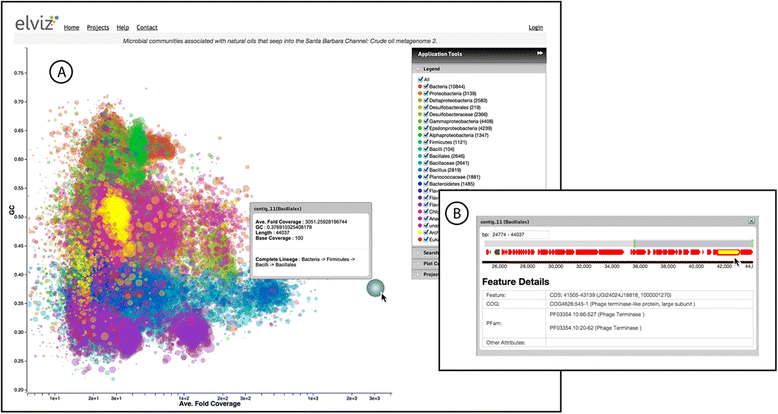
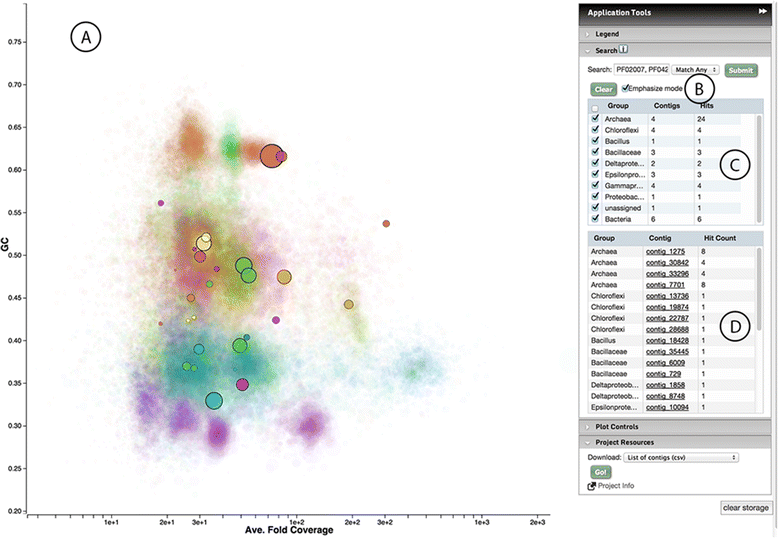
Results: Elviz (Environmental Laboratory Visualization) is an interactive web-based tool for the visual exploration of assembled metagenomes and their complex metadata. Elviz allows scientists to navigate metagenome assemblies across multiple dimensions and scales, plotting parameters such as GC content, relative abundance, phylogenetic affiliation and assembled contig length. Furthermore Elviz enables interactive exploration using real-time plot navigation, search, filters, axis selection, and the ability to drill from a whole-community profile down to individual gene annotations. Thus scientists engage in a rapid feedback loop of visual pattern identification, hypothesis generation, and hypothesis testing.
Conclusions: Compared to the current alternative of generating a succession of static figures, Elviz can greatly accelerate the speed of metagenome analysis. Elviz can be used to explore both user-submitted datasets and numerous metagenome studies publicly available at the Joint Genome Institute (JGI). Elviz is freely available at http://genome.jgi.doe.gov/viz and runs on most current web-browsers.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

