Systems biology of host-microbe metabolomics
- PMID: 25929487
- PMCID: PMC5029777
- DOI: 10.1002/wsbm.1301
Systems biology of host-microbe metabolomics
Abstract
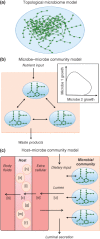
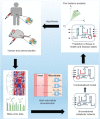
The human gut microbiota performs essential functions for host and well-being, but has also been linked to a variety of disease states, e.g., obesity and type 2 diabetes. The mammalian body fluid and tissue metabolomes are greatly influenced by the microbiota, with many health-relevant metabolites being considered 'mammalian-microbial co-metabolites'. To systematically investigate this complex host-microbial co-metabolism, a systems biology approach integrating high-throughput data and computational network models is required. Here, we review established top-down and bottom-up systems biology approaches that have successfully elucidated relationships between gut microbiota-derived metabolites and host health and disease. We focus particularly on the constraint-based modeling and analysis approach, which enables the prediction of mechanisms behind metabolic host-microbe interactions on the molecular level. We illustrate that constraint-based models are a useful tool for the contextualization of metabolomic measurements and can further our insight into host-microbe interactions, yielding, e.g., in potential novel drugs and biomarkers.
© 2015 The Authors. WIREs Systems Biology and Medicine published by Wiley Periodicals, Inc.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

