Lineage-specific sequence evolution and exon edge conservation partially explain the relationship between evolutionary rate and expression level in A. thaliana
- PMID: 25930165
- PMCID: PMC4480654
- DOI: 10.1111/mec.13221
Lineage-specific sequence evolution and exon edge conservation partially explain the relationship between evolutionary rate and expression level in A. thaliana
Abstract
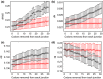
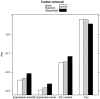
Rapidly evolving proteins can aid the identification of genes underlying phenotypic adaptation across taxa, but functional and structural elements of genes can also affect evolutionary rates. In plants, the 'edges' of exons, flanking intron junctions, are known to contain splice enhancers and to have a higher degree of conservation compared to the remainder of the coding region. However, the extent to which these regions may be masking indicators of positive selection or account for the relationship between dN/dS and other genomic parameters is unclear. We investigate the effects of exon edge conservation on the relationship of dN/dS to various sequence characteristics and gene expression parameters in the model plant Arabidopsis thaliana. We also obtain lineage-specific dN/dS estimates, making use of the recently sequenced genome of Thellungiella parvula, the second closest sequenced relative after the sister species Arabidopsis lyrata. Overall, we find that the effect of exon edge conservation, as well as the use of lineage-specific substitution estimates, upon dN/dS ratios partly explains the relationship between the rates of protein evolution and expression level. Furthermore, the removal of exon edges shifts dN/dS estimates upwards, increasing the proportion of genes potentially under adaptive selection. We conclude that lineage-specific substitutions and exon edge conservation have an important effect on dN/dS ratios and should be considered when assessing their relationship with other genomic parameters.
Keywords: Arabidopsis thaliana; dN/dS; lineage-specific evolution; splice enhancer.
© 2015 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures
References
-
- Akashi H, Eyre-Walker A. Translational selection and molecular evolution. Current Opinion in Genetics & Development. 1998;8:688–693. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. - PubMed
-
- Baerenfaller K, Grossmann J, Grobei MA, et al. Genome-scale proteomics reveals arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

