An R2R3-MYB Transcription Factor Regulates Eugenol Production in Ripe Strawberry Fruit Receptacles
- PMID: 25931522
- PMCID: PMC4453772
- DOI: 10.1104/pp.114.252908
An R2R3-MYB Transcription Factor Regulates Eugenol Production in Ripe Strawberry Fruit Receptacles
Abstract
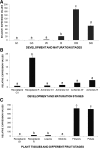
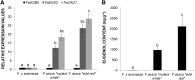
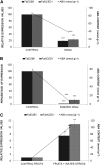
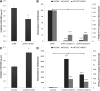
Eugenol is a volatile phenylpropanoid that contributes to flower and ripe fruit scent. In ripe strawberry (Fragaria × ananassa) fruit receptacles, eugenol is biosynthesized by eugenol synthase (FaEGS2). However, the transcriptional regulation of this process is still unknown. We have identified and functionally characterized an R2R3 MYB transcription factor (emission of benzenoid II [FaEOBII]) that seems to be the orthologous gene of PhEOBII from Petunia hybrida, which contributes to the regulation of eugenol biosynthesis in petals. The expression of FaEOBII was ripening related and fruit receptacle specific, although high expression values were also found in petals. This expression pattern of FaEOBII correlated with eugenol content in both fruit receptacle and petals. The expression of FaEOBII was repressed by auxins and activated by abscisic acid, in parallel to the ripening process. In ripe strawberry receptacles, where the expression of FaEOBII was silenced, the expression of cinnamyl alcohol dehydrogenase1 and FaEGS2, two structural genes involved in eugenol production, was down-regulated. A subsequent decrease in eugenol content in ripe receptacles was also observed, confirming the involvement of FaEOBII in eugenol metabolism. Additionally, the expression of FaEOBII was under the control of FaMYB10, another R2R3 MYB transcription factor that regulates the early and late biosynthetic genes from the flavonoid/phenylpropanoid pathway. In parallel, the amount of eugenol in FaMYB10-silenced receptacles was also diminished. Taken together, these data indicate that FaEOBII plays a regulating role in the volatile phenylpropanoid pathway gene expression that gives rise to eugenol production in ripe strawberry receptacles.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures






References
-
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 - PubMed
-
- Asif M, Dhawan P, Nath P (2000) A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol Biol Rep 18: 109–115
-
- Aubert C, Pitrat M (2006) Volatile compounds in the skin and pulp of Queen Anne’s pocket melon. J Agric Food Chem 54: 8177–8182 - PubMed
-
- Benítez-Burraco A, Blanco-Portales R, Redondo-Nevado J, Bellido ML, Moyano E, Caballero JL, Muñoz-Blanco J (2003) Cloning and characterization of two ripening-related strawberry (Fragaria × ananassa cv. Chandler) pectate lyase genes. J Exp Bot 54: 633–645 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

