The evolutionary fate of alternatively spliced homologous exons after gene duplication
- PMID: 25931610
- PMCID: PMC4494069
- DOI: 10.1093/gbe/evv076
The evolutionary fate of alternatively spliced homologous exons after gene duplication
Abstract
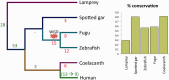
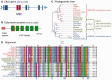
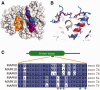
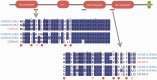
Alternative splicing and gene duplication are the two main processes responsible for expanding protein functional diversity. Although gene duplication can generate new genes and alternative splicing can introduce variation through alternative gene products, the interplay between the two processes is complex and poorly understood. Here, we have carried out a study of the evolution of alternatively spliced exons after gene duplication to better understand the interaction between the two processes. We created a manually curated set of 97 human genes with mutually exclusively spliced homologous exons and analyzed the evolution of these exons across five distantly related vertebrates (lamprey, spotted gar, zebrafish, fugu, and coelacanth). Most of these exons had an ancient origin (more than 400 Ma). We found examples supporting two extreme evolutionary models for the behaviour of homologous axons after gene duplication. We observed 11 events in which gene duplication was accompanied by splice isoform separation, that is, each paralog specifically conserved just one distinct ancestral homologous exon. At other extreme, we identified genes in which the homologous exons were always conserved within paralogs, suggesting that the alternative splicing event cannot easily be separated from the function in these genes. That many homologous exons fall in between these two extremes highlights the diversity of biological systems and suggests that the subtle balance between alternative splicing and gene duplication is adjusted to the specific cellular context of each gene.
Keywords: alternative splicing; gene duplication; homologous exons; protein diversity; subfunctionalization.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Evidence for widespread subfunctionalization of splice forms in vertebrate genomes.Genome Res. 2015 May;25(5):624-32. doi: 10.1101/gr.184473.114. Epub 2015 Mar 19. Genome Res. 2015. PMID: 25792610 Free PMC article.
-
Alternatively Spliced Homologous Exons Have Ancient Origins and Are Highly Expressed at the Protein Level.PLoS Comput Biol. 2015 Jun 10;11(6):e1004325. doi: 10.1371/journal.pcbi.1004325. eCollection 2015 Jun. PLoS Comput Biol. 2015. PMID: 26061177 Free PMC article.
-
Gene duplication followed by exon structure divergence substitutes for alternative splicing in zebrafish.Gene. 2014 Aug 10;546(2):271-6. doi: 10.1016/j.gene.2014.05.068. Epub 2014 Jun 3. Gene. 2014. PMID: 24942242
-
Gene Duplication and Alternative Splicing as Evolutionary Drivers of Proteome Specialization.Bioessays. 2025 May;47(5):e202400202. doi: 10.1002/bies.202400202. Epub 2025 Feb 25. Bioessays. 2025. PMID: 39995355 Review.
-
Evolutionary convergence of alternative splicing in ion channels.Trends Genet. 2004 Apr;20(4):171-6. doi: 10.1016/j.tig.2004.02.001. Trends Genet. 2004. PMID: 15101391 Review.
Cited by
-
Toward a comprehensive profiling of alternative splicing proteoform structures, interactions and functions.Curr Opin Struct Biol. 2025 Feb;90:102979. doi: 10.1016/j.sbi.2024.102979. Epub 2025 Jan 7. Curr Opin Struct Biol. 2025. PMID: 39778413 Free PMC article. Review.
-
Origins and Evolution of Human Tandem Duplicated Exon Substitution Events.Genome Biol Evol. 2022 Dec 7;14(12):evac162. doi: 10.1093/gbe/evac162. Genome Biol Evol. 2022. PMID: 36346145 Free PMC article.
-
The Rho GTPase Family Genes in Bivalvia Genomes: Sequence, Evolution and Expression Analysis.PLoS One. 2015 Dec 3;10(12):e0143932. doi: 10.1371/journal.pone.0143932. eCollection 2015. PLoS One. 2015. PMID: 26633655 Free PMC article.
-
The divergence of alternative splicing between ohnologs in teleost fishes.BMC Ecol Evol. 2021 May 25;21(1):98. doi: 10.1186/s12862-021-01833-6. BMC Ecol Evol. 2021. PMID: 34034651 Free PMC article.
-
The ribosome-engaged landscape of alternative splicing.Nat Struct Mol Biol. 2016 Dec;23(12):1117-1123. doi: 10.1038/nsmb.3317. Epub 2016 Nov 7. Nat Struct Mol Biol. 2016. PMID: 27820807 Free PMC article.
References
-
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

