Cu(2+)-RGDFRGDS: exploring the mechanism and high efficacy of the nanoparticle in antithrombotic therapy
- PMID: 25931819
- PMCID: PMC4404989
- DOI: 10.2147/IJN.S76691
Cu(2+)-RGDFRGDS: exploring the mechanism and high efficacy of the nanoparticle in antithrombotic therapy
Abstract
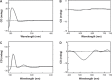
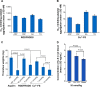
Thrombosis disease has been the leading cause of morbidity and mortality worldwide. In the discovery of antithrombotic agents, three complexes of Cu(2+) and repetitive arginine-glycine-aspartic acid (RGD) sequences, Cu(II)-Arg-Gly-Asp-Ser-Arg-Gly-Asp-Ser (Cu[II]-4a), Cu(II)-Arg-Gly-Asp-Val-Arg-Gly-Asp-Val (Cu[II]-4b), and Cu(II)-Arg-Gly-Asp-Phe-Arg-Gly-Asp-Phe (Cu[II]-4c), were previously reported, of which Cu(II)-4a and Cu(II)-4c possessed the highest in vitro and in vivo activity, respectively. Transmission electron microscopy (TEM) images visualized that Cu(II)-4a and Cu(II)-4c formed nanoaggregates and nanoparticles, respectively. However, the details of the formation of the nanospecies complexes and of the mechanism for inhibiting thrombosis remain to be clarified. For this purpose, this study designed a novel complex of Cu(II) and the RGD octapeptide, Arg-Gly-Asp-Phe-Arg-Gly-Asp-Ser (RGDFRGDS), consisting of Arg-Gly-Asp-Phe of Cu(II)-4c and Arg-Gly-Asp-Ser of Cu(II)-4a, to colligate their biological and nanostructural benefits. In contrast with Cu(II)-4a, -4b, and -4c, Cu(II)-RGDFRGDS (Cu(2+)-FS) had high antiplatelet and antithrombotic activities, with the formed nanoparticles having a porous surface. Additionally, this paper evidenced the dimer had the basic structural unit of Cu(2+)-FS in water, theoretically simulated the formation of Cu(2+)-FS nanoparticles, and identified that Cu(2+)-FS activity in decreasing glycoprotein IIb/IIIa, P-selectin, and IL-8 was responsible for the antithrombotic action. Finally, adherence onto the surface and entry into the cytoplasm were considered the steps of a two-step model for the blocking of platelet activation by Cu(2+)-FS nanoparticles. Findings indicated that the antiplatelet aggregation activity of Cu(2+)-FS was 10-52 times higher than that of RGDFRGDS, while the effective dose for antithrombotic action was 5,000 times lower than that of RGDFRGDS.
Keywords: AFM; GPIIb/IIIa; IL-8; SEM; TEM; nanomedicine.
Figures









Similar articles
-
Novel Cu(II)-RGD-octapeptides: Synthesis, coordination mode, in vitro anti-platelet aggregation/in vivo anti-thrombotic evaluation and correlation of sequence with nano-structure.Nanomedicine. 2011 Aug;7(4):403-9. doi: 10.1016/j.nano.2011.01.005. Epub 2011 Jan 25. Nanomedicine. 2011. PMID: 21272663
-
Nanosized aspirin-Arg-Gly-Asp-Val: delivery of aspirin to thrombus by the target carrier Arg-Gly-Asp-Val tetrapeptide.ACS Nano. 2013 Sep 24;7(9):7664-73. doi: 10.1021/nn402171v. Epub 2013 Aug 29. ACS Nano. 2013. PMID: 23931063
-
The application of tetrahydroisoquinoline-3-carbonyl-TARGD(F)F as an anti-thrombotic agent having dual mechanisms of action.Mol Biosyst. 2012 Oct;8(10):2672-9. doi: 10.1039/c2mb25112d. Mol Biosyst. 2012. PMID: 22801714
-
[A new thrombocyte aggregation-inhibiting and fibrinolysis-promoting synthetic molecule: RGDF (Arg-Gly-Asp-Phe) coupled with the carboxyterminal antiplasmin peptide].Orv Hetil. 1995 Jan 15;136(3):129-33. Orv Hetil. 1995. PMID: 7870410 Review. Hungarian.
-
Novel antithrombotic drugs in development.Drugs. 1995 Jun;49(6):856-84. doi: 10.2165/00003495-199549060-00002. Drugs. 1995. PMID: 7641602 Review.
Cited by
-
Dimethyl 2,2'-[2,2'-(ethane-1,1-diyl)bis(1H-indole-3,2-diyl)]-diacetate: a small molecule capable of nano-scale assembly, inhibiting venous thrombosis and inducing no bleeding side effect.Int J Nanomedicine. 2018 Nov 22;13:7835-7844. doi: 10.2147/IJN.S178683. eCollection 2018. Int J Nanomedicine. 2018. PMID: 30538462 Free PMC article.
-
Doxorubicin and anti-VEGF siRNA co-delivery via nano-graphene oxide for enhanced cancer therapy in vitro and in vivo.Int J Nanomedicine. 2018 Jun 27;13:3713-3728. doi: 10.2147/IJN.S162939. eCollection 2018. Int J Nanomedicine. 2018. PMID: 29983564 Free PMC article.
-
RGD(F/S/V)-Dex: towards the development of novel, effective, and safe glucocorticoids.Drug Des Devel Ther. 2016 Mar 8;10:1059-76. doi: 10.2147/DDDT.S99568. eCollection 2016. Drug Des Devel Ther. 2016. PMID: 27022245 Free PMC article.
-
Aqueous extract of Rabdosia rubescens leaves: forming nanoparticles, targeting P-selectin, and inhibiting thrombosis.Int J Nanomedicine. 2015 Nov 4;10:6905-18. doi: 10.2147/IJN.S91316. eCollection 2015. Int J Nanomedicine. 2015. PMID: 26604756 Free PMC article.
-
BCESA: a nano-scaled intercalator capable of targeting tumor tissue and releasing anti-tumoral β-carboline-3-carboxylic acid.Int J Nanomedicine. 2019 Apr 30;14:3027-3041. doi: 10.2147/IJN.S187600. eCollection 2019. Int J Nanomedicine. 2019. PMID: 31118620 Free PMC article.
References
-
- Arnaout MA, Goodman SL, Xiong JP. Coming to grips with integrin binding to ligands. Curr Opin Cell Biol. 2002;14(5):641–651. - PubMed
-
- Malkar NB, Lauer-Fields JL, Juska D, Fields GB. Characterization of peptide-amphiphiles possessing cellular activation sequences. Biomacromolecules. 2003;4(3):518–528. - PubMed
-
- Conradi J, Huber S, Gaus K, et al. Cyclic RGD peptides interfere with binding of the Helicobacter pylori protein CagL to integrins αVβ3 and α5β1. Amino Acids. 2012;43(1):219–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

