The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1
- PMID: 25933022
- PMCID: PMC4416901
- DOI: 10.1371/journal.pone.0125345
The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1
Erratum in
-
Correction: The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1.PLoS One. 2015 Jun 19;10(6):e0131132. doi: 10.1371/journal.pone.0131132. eCollection 2015. PLoS One. 2015. PMID: 26091017 Free PMC article. No abstract available.
Abstract
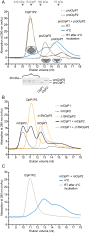
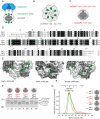
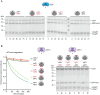
Clp chaperone-proteases are cylindrical complexes built from ATP-dependent chaperonerings that stack onto a proteolytic ClpP double-ring core to carry out substrate protein degradation.Interaction of the ClpP particle with the chaperone is mediated by an N-terminal loop and a hydrophobic surface patch on the ClpP ring surface. In contrast to E. coli, Myco bacterium tuberculosis harbors not only one but two ClpP protease subunits, ClpP1 and ClpP2,and a homo-heptameric ring of each assembles to form the ClpP1P2 double-ring core. Consequently,this hetero double-ring presents two different potential binding surfaces for the interaction with the chaperones ClpX and ClpC1. To investigate whether ClpX or ClpC1 might preferentially interact with one or the other double-ring face, we mutated the hydrophobicchaperone-interaction patch on either ClpP1 or ClpP2, generating ClpP1P2 particles that are defective in one of the two binding patches and thereby in their ability to interact with their chaperone partners. Using chaperone-mediated degradation of ssrA-tagged model substrates, we show that both Mycobacterium tuberculosis Clp chaperones require the intact interaction face of ClpP2 to support degradation, resulting in an asymmetric complex where chaperones only bind to the ClpP2 side of the proteolytic core. This sets the Clpproteases of Mycobacterium tuberculosis, and probably other Actinobacteria, apart from the well-studied E. coli system, where chaperones bind to both sides of the protease core,and it frees the ClpP1 interaction interface for putative new binding partners [corrected].
Conflict of interest statement
Figures

Similar articles
-
Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase.Nat Struct Biol. 2001 Mar;8(3):230-3. doi: 10.1038/84967. Nat Struct Biol. 2001. PMID: 11224567
-
Specificity in substrate and cofactor recognition by the N-terminal domain of the chaperone ClpX.Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17724-9. doi: 10.1073/pnas.0601505103. Epub 2006 Nov 7. Proc Natl Acad Sci U S A. 2006. PMID: 17090685 Free PMC article.
-
Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis.J Biol Chem. 2001 Aug 3;276(31):29420-9. doi: 10.1074/jbc.M103489200. Epub 2001 May 9. J Biol Chem. 2001. PMID: 11346657
-
[Bacterial ClpX protease structure and function--a review].Wei Sheng Wu Xue Bao. 2010 Oct;50(10):1281-7. Wei Sheng Wu Xue Bao. 2010. PMID: 21141460 Review. Chinese.
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
Cited by
-
Identification of a phenyl ester covalent inhibitor of caseinolytic protease and analysis of the ClpP1P2 inhibition in mycobacteria.mLife. 2025 Apr 15;4(2):155-168. doi: 10.1002/mlf2.12169. eCollection 2025 Apr. mLife. 2025. PMID: 40313980 Free PMC article.
-
Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.J Biol Chem. 2016 Apr 1;291(14):7465-76. doi: 10.1074/jbc.M115.700344. Epub 2016 Feb 8. J Biol Chem. 2016. PMID: 26858247 Free PMC article.
-
Acyldepsipeptide Analogues: A Future Generation Antibiotics for Tuberculosis Treatment.Pharmaceutics. 2022 Sep 15;14(9):1956. doi: 10.3390/pharmaceutics14091956. Pharmaceutics. 2022. PMID: 36145704 Free PMC article. Review.
-
Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides.Chem Rev. 2022 Sep 28;122(18):14722-14814. doi: 10.1021/acs.chemrev.2c00210. Epub 2022 Sep 1. Chem Rev. 2022. PMID: 36049139 Free PMC article. Review.
-
Cellular functions of the ClpP protease impacting bacterial virulence.Front Mol Biosci. 2022 Dec 1;9:1054408. doi: 10.3389/fmolb.2022.1054408. eCollection 2022. Front Mol Biosci. 2022. PMID: 36533084 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

