Epstein-Barr virus genetic variation in lymphoblastoid cell lines derived from Kenyan pediatric population
- PMID: 25933165
- PMCID: PMC4416826
- DOI: 10.1371/journal.pone.0125420
Epstein-Barr virus genetic variation in lymphoblastoid cell lines derived from Kenyan pediatric population
Erratum in
-
Correction: epstein-barr virus genetic variation in lymphoblastoid cell lines derived from Kenyan pediatric population.PLoS One. 2015 Jun 3;10(6):e0130229. doi: 10.1371/journal.pone.0130229. eCollection 2015. PLoS One. 2015. PMID: 26039342 Free PMC article. No abstract available.
Abstract
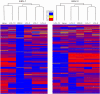
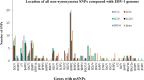
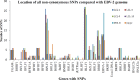
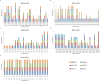
Epstein-Barr virus (EBV) is associated with Burkitt's lymphoma (BL), and in regions of sub-Saharan Africa where endemic BL is common, both the EBV Type 1 (EBV-1) and EBV Type 2 strains (EBV-2) are found. Little is known about genetic variation of EBV strains in areas of sub-Saharan Africa. In the present study, spontaneous lymphoblastoid cell lines (LCLs) were generated from samples obtained from Kenya. Polymerase chain reaction (PCR) amplification of the EBV genome was done using multiple primers and sequenced by next-generation sequencing (NGS). Phylogenetic analyses against the published EBV-1 and EBV-2 strains indicated that one sample, LCL10 was closely related to EBV-2, while the remaining 3 LCL samples were more closely related to EBV-1. Moreover, single nucleotide polymorphism (SNP) analyses showed clustering of LCL variants. We further show by analysis of EBNA-1, BLLF1, BPLF1, and BRRF2 that latent genes are less conserved than lytic genes in these LCLs from a single geographic region. In this study we have shown that NGS is highly useful for deciphering detailed inter and intra-variations in EBV genomes and that within a geographic region different EBV genetic variations can co-exist, the implications of which warrant further investigation. The findings will enhance our understanding of potential pathogenic variants critical to the development and maintenance of EBV-associated malignancies.
Conflict of interest statement
Figures

Similar articles
-
Epstein-Barr Virus Genomes Reveal Population Structure and Type 1 Association with Endemic Burkitt Lymphoma.J Virol. 2020 Aug 17;94(17):e02007-19. doi: 10.1128/JVI.02007-19. Print 2020 Aug 17. J Virol. 2020. PMID: 32581102 Free PMC article.
-
Epstein-Barr virus from Burkitt Lymphoma biopsies from Africa and South America share novel LMP-1 promoter and gene variations.Sci Rep. 2015 Nov 23;5:16706. doi: 10.1038/srep16706. Sci Rep. 2015. PMID: 26593963 Free PMC article.
-
RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes.J Virol. 2019 Jun 14;93(13):e00226-19. doi: 10.1128/JVI.00226-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019051 Free PMC article.
-
The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: a need for reappraisal.Virus Res. 2009 Aug;143(2):209-21. doi: 10.1016/j.virusres.2009.07.005. Epub 2009 Jul 23. Virus Res. 2009. PMID: 19596032 Free PMC article. Review.
-
Phylogenetic comparison of Epstein-Barr virus genomes.J Microbiol. 2018 Aug;56(8):525-533. doi: 10.1007/s12275-018-8039-x. Epub 2018 Jun 14. J Microbiol. 2018. PMID: 29948828 Review.
Cited by
-
From Conventional to Next Generation Sequencing of Epstein-Barr Virus Genomes.Viruses. 2016 Feb 24;8(3):60. doi: 10.3390/v8030060. Viruses. 2016. PMID: 26927157 Free PMC article. Review.
-
A Review of Cancer Genetics and Genomics Studies in Africa.Front Oncol. 2021 Feb 15;10:606400. doi: 10.3389/fonc.2020.606400. eCollection 2020. Front Oncol. 2021. PMID: 33659210 Free PMC article. Review.
-
Molecular Characterisation of Epstein-Barr Virus in Classical Hodgkin Lymphoma.Int J Mol Sci. 2022 Dec 9;23(24):15635. doi: 10.3390/ijms232415635. Int J Mol Sci. 2022. PMID: 36555277 Free PMC article.
-
Epstein-Barr Virus Type 2 Infects T Cells and Induces B Cell Lymphomagenesis in Humanized Mice.J Virol. 2018 Oct 12;92(21):e00813-18. doi: 10.1128/JVI.00813-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30089703 Free PMC article.
-
Evidence of EBV infection in lymphomas diagnosed in Lusaka, Zambia.Pan Afr Med J. 2018 Mar 28;29:181. doi: 10.11604/pamj.2018.29.181.11847. eCollection 2018. Pan Afr Med J. 2018. PMID: 30061959 Free PMC article.
References
-
- Kutok JL, Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol. 2006;1:375–404. - PubMed
-
- McGeoch DJ, Gatherer D. Lineage structures in the genome sequences of three Epstein-Barr virus strains. Virology. 2007. March 1;359(1):1–5. Epub 2006 Nov 13. - PubMed
-
- Edwards RH, Seillier-Moiseiwitsch F, Raab-Traub N. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology. 1999. August 15;261(1):79–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

