Differentiation of IncL and IncM Plasmids Associated with the Spread of Clinically Relevant Antimicrobial Resistance
- PMID: 25933288
- PMCID: PMC4416936
- DOI: 10.1371/journal.pone.0123063
Differentiation of IncL and IncM Plasmids Associated with the Spread of Clinically Relevant Antimicrobial Resistance
Abstract
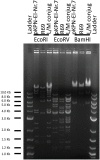
Introduction: blaOXA-48, blaNDM-1 and blaCTX-M-3 are clinically relevant resistance genes, frequently associated with the broad-host range plasmids of the IncL/M group. The L and M plasmids belong to two compatible groups, which were incorrectly classified together by molecular methods. In order to understand their evolution, we fully sequenced four IncL/M plasmids, including the reference plasmids R471 and R69, the recently described blaOXA-48-carrying plasmid pKPN-El.Nr7 from a Klebsiella pneumoniae isolated in Bern (Switzerland), and the blaSHV-5 carrying plasmid p202c from a Salmonella enterica from Tirana (Albania).
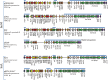
Methods: Sequencing was performed using 454 Junior Genome Sequencer (Roche). Annotation was performed using Sequin and Artemis software. Plasmid sequences were compared with 13 fully sequenced plasmids belonging to the IncL/M group available in GenBank.
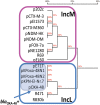
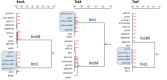
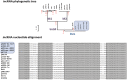
Results: Comparative analysis of plasmid genomes revealed two distinct genetic lineages, each containing one of the R471 (IncL) and R69 (IncM) reference plasmids. Conjugation experiments demonstrated that plasmids representative of the IncL and IncM groups were compatible with each other. The IncL group is constituted by the blaOXA-48-carrying plasmids and R471. The IncM group contains two sub-types of plasmids named IncM1 and IncM2 that are each incompatible.
Conclusion: This work re-defines the structure of the IncL and IncM families and ascribes a definitive designation to the fully sequenced IncL/M plasmids available in GenBank.
Conflict of interest statement
Figures
References
-
- Smet A, Van Nieuwerburgh F, Vandekerckhove TT, Martel A, Deforce D, Butaye P, et al. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLoS One 2010;5: e11202 10.1371/journal.pone.0011202 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

