Protein Degradation of RNA Polymerase II-Association Factor 1(PAF1) Is Controlled by CNOT4 and 26S Proteasome
- PMID: 25933433
- PMCID: PMC4416890
- DOI: 10.1371/journal.pone.0125599
Protein Degradation of RNA Polymerase II-Association Factor 1(PAF1) Is Controlled by CNOT4 and 26S Proteasome
Abstract
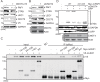
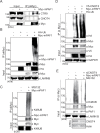
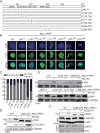
The PAF complex (PAFc) participates in various steps of the transcriptional process, from initiation to termination, by interacting with and recruiting various proteins to the proper locus for each step. PAFc is an evolutionarily conserved, multi-protein complex comprising PAF1, CDC73, CTR9, LEO1, yRTF1 and, in humans, hSKI8. These components of PAFc work together, and their protein levels are closely interrelated. In the present study, we investigated the mechanism of PAF1 protein degradation. We found that PAF1 protein levels are negatively regulated by the expression of CNOT4, an ortholog of yNOT4 and a member of the CCR4-NOT complex. CNOT4 specifically controls PAF1 but not other components of PAFc at the protein level by regulating the polyubiquitination of PAF1 and its subsequent degradation by the 26S proteasome. The degradation of PAF1 was found to require nuclear localization, as no PAF1 degradation by CNOT4 and the 26S proteasome was observed with NLS (nucleus localization signal)-deficient PAF1 mutants. However, chromatin binding by PAF1 was not necessary for 26S proteasome- or CNOT4-mediated degradation. Our results suggest that CNOT4 controls the degradation of chromatin-unbound PAF1 via the 26S proteasome.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

