The methyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex
- PMID: 25934499
- PMCID: PMC4432814
- DOI: 10.1186/s13064-015-0040-z
The methyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex
Abstract
Background: Chromatin-modifying complexes have key roles in regulating various aspects of neural stem cell biology, including self-renewal and neurogenesis. The methyl binding domain 3/nucleosome remodelling and deacetylation (MBD3/NuRD) co-repressor complex facilitates lineage commitment of pluripotent cells in early mouse embryos and is important for stem cell homeostasis in blood and skin, but its function in neurogenesis had not been described. Here, we show for the first time that MBD3/NuRD function is essential for normal neurogenesis in mice.
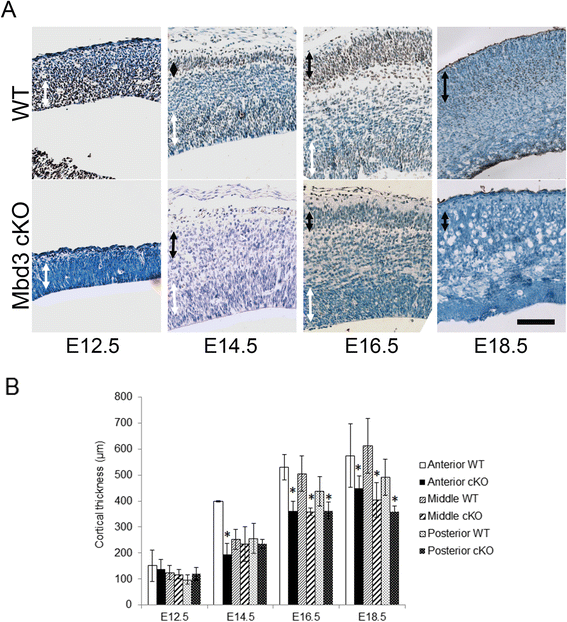
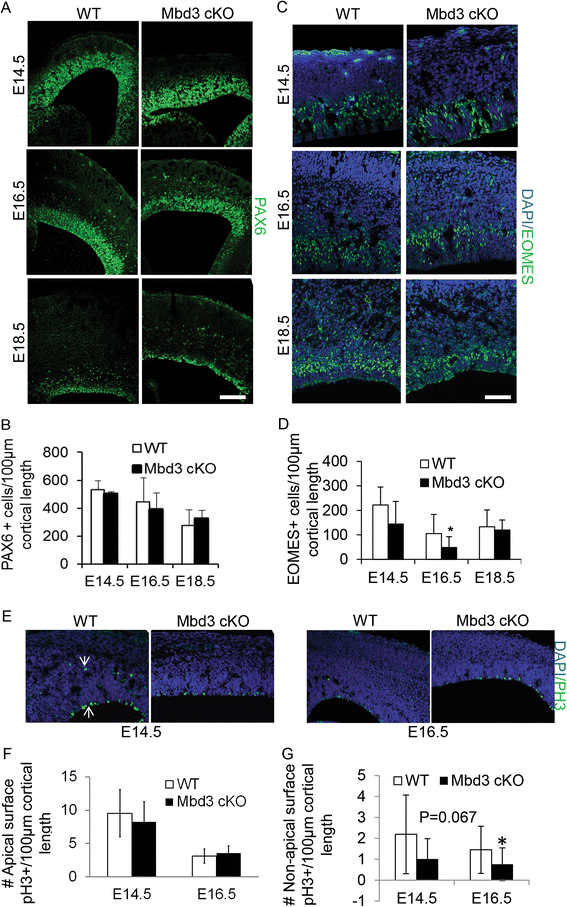
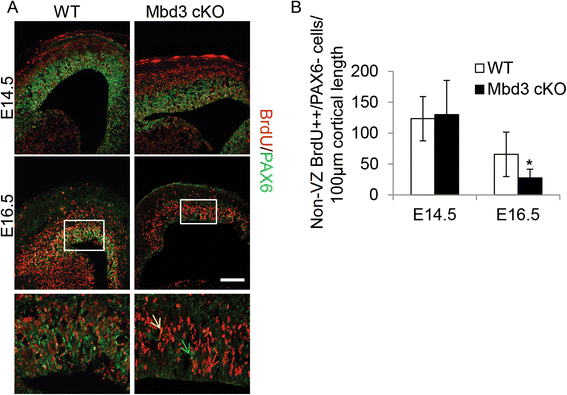
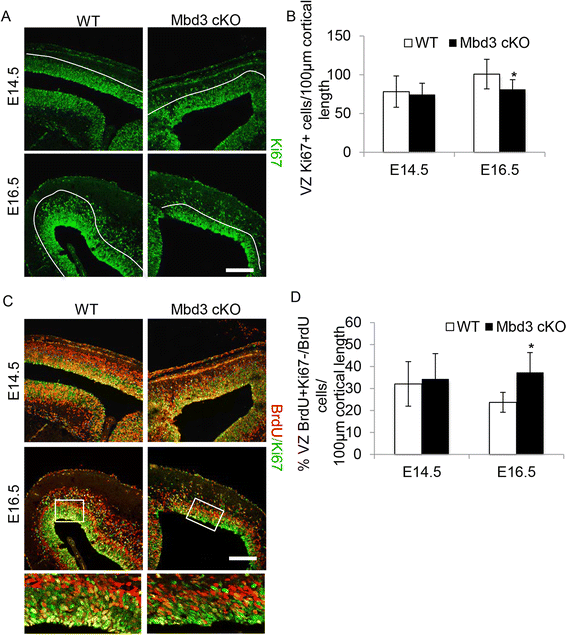
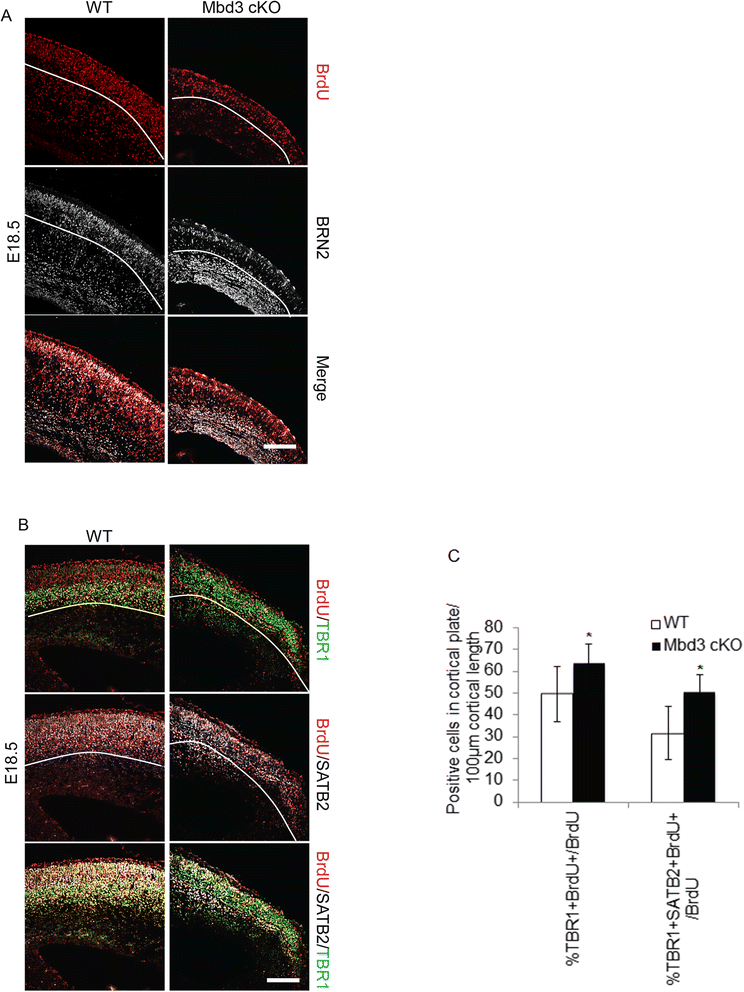
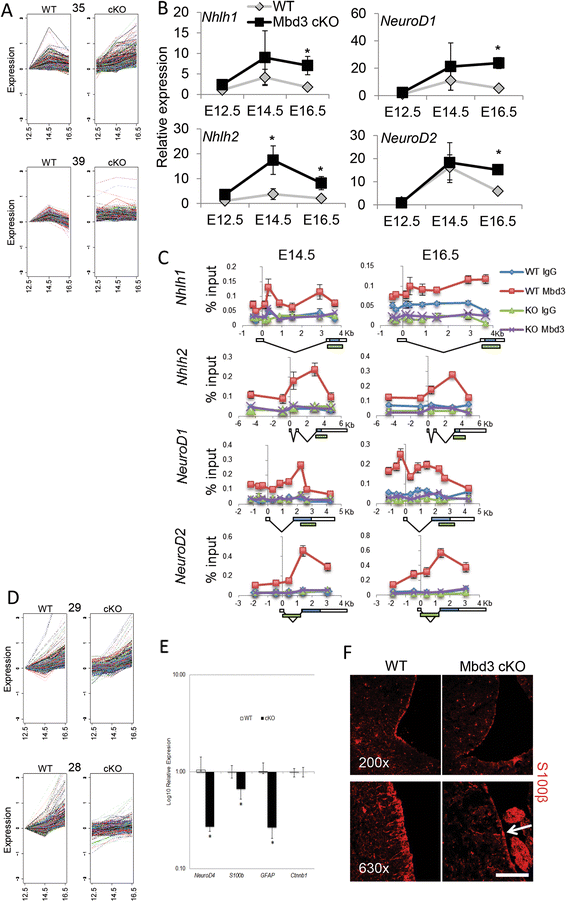
Results: Deletion of MBD3, a structural component of the NuRD complex, in the developing mouse central nervous system resulted in reduced cortical thickness, defects in the proper specification of cortical projection neuron subtypes and neonatal lethality. These phenotypes are due to alterations in PAX6+ apical progenitor cell outputs, as well as aberrant terminal neuronal differentiation programmes of cortical plate neurons. Normal numbers of PAX6+ apical neural progenitor cells were generated in the MBD3/NuRD-mutant cortex; however, the PAX6+ apical progenitor cells generate EOMES+ basal progenitor cells in reduced numbers. Cortical progenitor cells lacking MBD3/NuRD activity generate neurons that express both deep- and upper-layer markers. Using laser capture microdissection, gene expression profiling and chromatin immunoprecipitation, we provide evidence that MBD3/NuRD functions to control gene expression patterns during neural development.
Conclusions: Our data suggest that although MBD3/NuRD is not required for neural stem cell lineage commitment, it is required to repress inappropriate transcription in both progenitor cells and neurons to facilitate appropriate cell lineage choice and differentiation programmes.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

