The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site
- PMID: 25934501
- PMCID: PMC4421977
- DOI: 10.1101/gad.261974.115
The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site
Abstract
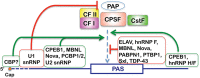
The key RNA sequence elements and protein factors necessary for 3' processing of polyadenylated mRNA precursors are well known. Recent studies, however, have significantly reshaped current models for the protein-RNA interactions involved in poly(A) site recognition, painting a picture more complex than previously envisioned and also providing new insights into regulation of this important step in gene expression. Here we review the recent advances in this area and provide a perspective for future studies.
Keywords: gene expression; mRNA processing; poly(A) site recognition.
© 2015 Shi and Manley; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell 25: 863–875. - PubMed
-
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11: 1703–1716. - PubMed
-
- Bava FA, Eliscovich C, Ferreira PG, Minana B, Ben-Dov C, Guigo R, Valcarcel J, Mendez R. 2013. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495: 121–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
