A chromatin code for alternative splicing involving a putative association between CTCF and HP1α proteins
- PMID: 25934638
- PMCID: PMC4446157
- DOI: 10.1186/s12915-015-0141-5
A chromatin code for alternative splicing involving a putative association between CTCF and HP1α proteins
Abstract
Background: Alternative splicing is primarily controlled by the activity of splicing factors and by the elongation of the RNA polymerase II (RNAPII). Recent experiments have suggested a new complex network of splicing regulation involving chromatin, transcription and multiple protein factors. In particular, the CCCTC-binding factor (CTCF), the Argonaute protein AGO1, and members of the heterochromatin protein 1 (HP1) family have been implicated in the regulation of splicing associated with chromatin and the elongation of RNAPII. These results raise the question of whether these proteins may associate at the chromatin level to modulate alternative splicing.
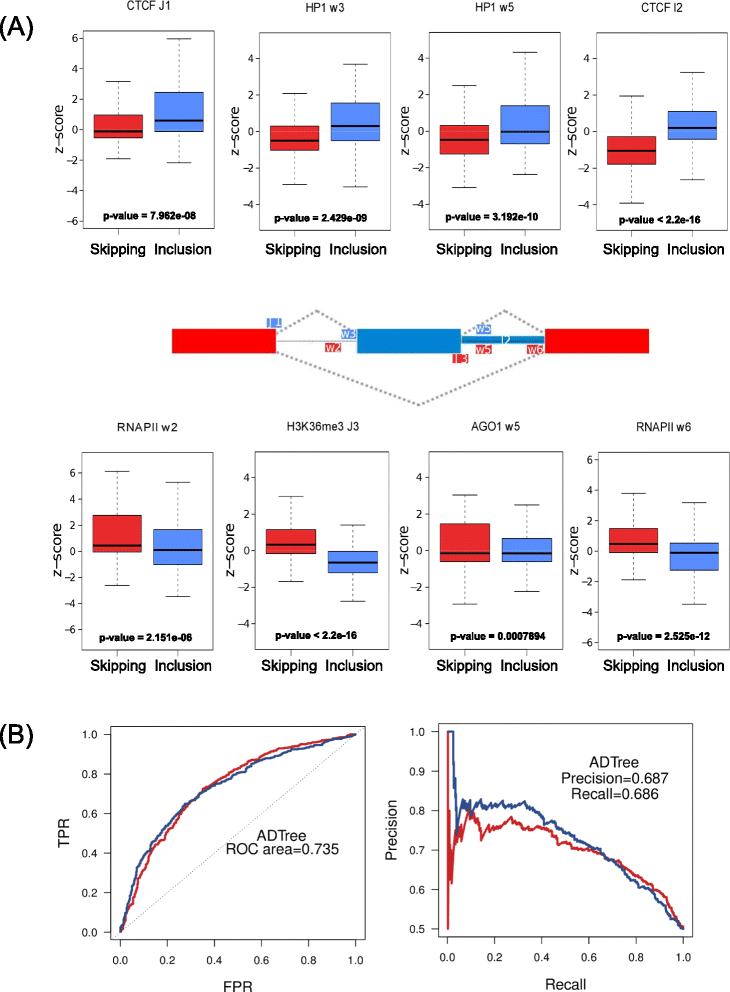
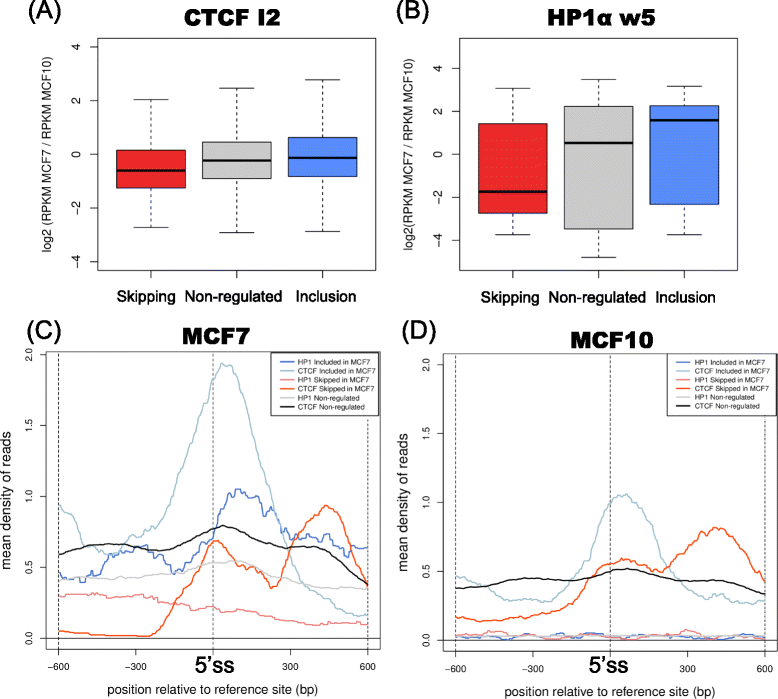
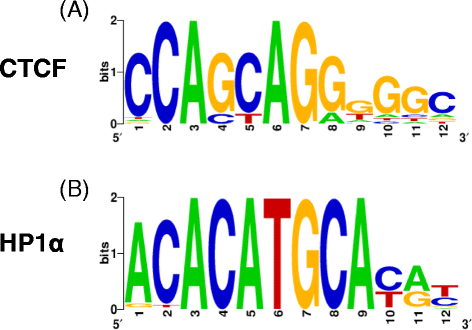
Results: Using chromatin immunoprecipitation sequencing (ChIP-Seq) data for CTCF, AGO1, HP1α, H3K27me3, H3K9me2, H3K36me3, RNAPII, total H3 and 5metC and alternative splicing arrays from two cell lines, we have analyzed the combinatorial code of their binding to chromatin in relation to the alternative splicing patterns between two cell lines, MCF7 and MCF10. Using Machine Learning techniques, we identified the changes in chromatin signals that are most significantly associated with splicing regulation between these two cell lines. Moreover, we have built a map of the chromatin signals on the pre-mRNA, that is, a chromatin-based RNA-map, which can explain 606 (68.55%) of the regulated events between MCF7 and MCF10. This chromatin code involves the presence of HP1α, CTCF, AGO1, RNAPII and histone marks around regulated exons and can differentiate patterns of skipping and inclusion. Additionally, we found a significant association of HP1α and CTCF activities around the regulated exons and a putative DNA binding site for HP1α.
Conclusions: Our results show that a considerable number of alternative splicing events could have a chromatin-dependent regulation involving the association of HP1α and CTCF near regulated exons. Additionally, we find further evidence for the involvement of HP1α and AGO1 in chromatin-related splicing regulation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

