The Mechanism and Function of Group II Chaperonins
- PMID: 25936650
- PMCID: PMC4706738
- DOI: 10.1016/j.jmb.2015.04.013
The Mechanism and Function of Group II Chaperonins
Abstract
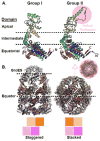
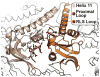
Protein folding in the cell requires the assistance of enzymes collectively called chaperones. Among these, the chaperonins are 1-MDa ring-shaped oligomeric complexes that bind unfolded polypeptides and promote their folding within an isolated chamber in an ATP-dependent manner. Group II chaperonins, found in archaea and eukaryotes, contain a built-in lid that opens and closes over the central chamber. In eukaryotes, the chaperonin TRiC/CCT is hetero-oligomeric, consisting of two stacked rings of eight paralogous subunits each. TRiC facilitates folding of approximately 10% of the eukaryotic proteome, including many cytoskeletal components and cell cycle regulators. Folding of many cellular substrates of TRiC cannot be assisted by any other chaperone. A complete structural and mechanistic understanding of this highly conserved and essential chaperonin remains elusive. However, recent work is beginning to shed light on key aspects of chaperonin function and how their unique properties underlie their contribution to maintaining cellular proteostasis.
Keywords: TRiC/CCT; chaperones; chaperonin; protein folding; proteostasis.
Copyright © 2015. Published by Elsevier Ltd.
Figures



References
-
- Levinthal C. How To Fold Graciously. University of Illinois Bulletin. 1969;67:22–4.
-
- Baldwin RL. The search for folding intermediates and the mechanism of protein folding. Annual review of biophysics. 2008;37:1–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

