Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics
- PMID: 25937169
- PMCID: PMC4551425
- DOI: 10.1016/j.neuron.2015.03.060
Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics
Abstract
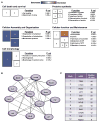
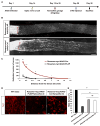
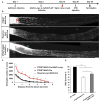
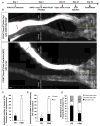
Neurons differ in their responses to injury, but the underlying mechanisms remain poorly understood. Using quantitative proteomics, we characterized the injury-triggered response from purified intact and axotomized retinal ganglion cells (RGCs). Subsequent informatics analyses revealed a network of injury-response signaling hubs. In addition to confirming known players, such as mTOR, this also identified new candidates, such as c-myc, NFκB, and Huntingtin. Similar to mTOR, c-myc has been implicated as a key regulator of anabolic metabolism and is downregulated by axotomy. Forced expression of c-myc in RGCs, either before or after injury, promotes dramatic RGC survival and axon regeneration after optic nerve injury. Finally, in contrast to RGCs, neither c-myc nor mTOR was downregulated in injured peripheral sensory neurons. Our studies suggest that c-myc and other injury-responsive pathways are critical to the intrinsic regenerative mechanisms and might represent a novel target for developing neural repair strategies in adults.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Bareyre FM, Kerschensteiner M, Raineteau O, Mettenleiter TC, Weinmann O, Schwab ME. The injured spinal cord spontaneously forms a new intraspinal circuit in adult rats. Nat Neurosci. 2004;7:269–277. - PubMed
-
- Bradke F, Fawcett JW, Spira ME. Assembly of a new growth cone after axotomy: the precursor to axon regeneration. Nat Rev Neurosci. 2012;13:183–193. - PubMed
-
- Chen A, Springer JE. Neuroproteomic methods in spinal cord injury. Methods Mol Biol. 2009;566:57–67. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

