Determination of hepatitis delta virus ribozyme N(-1) nucleobase and functional group specificity using internal competition kinetics
- PMID: 25937290
- PMCID: PMC4461535
- DOI: 10.1016/j.ab.2015.04.024
Determination of hepatitis delta virus ribozyme N(-1) nucleobase and functional group specificity using internal competition kinetics
Abstract
Biological catalysis involves interactions distant from the site of chemistry that can position the substrate for reaction. Catalysis of RNA 2'-O-transphosphorylation by the hepatitis delta virus (HDV) ribozyme is sensitive to the identity of the N(-1) nucleotide flanking the reactive phosphoryl group. However, the interactions that affect the conformation of this position, and in turn the 2'O nucleophile, are unclear. Here, we describe the application of multiple substrate internal competition kinetic analyses to understand how the N(-1) nucleobase contributes to HDV catalysis and test the utility of this approach for RNA structure-function studies. Internal competition reactions containing all four substrate sequence variants at the N(-1) position in reactions using ribozyme active site mutations at A77 and A78 were used to test a proposed base-pairing interaction. Mutants A78U, A78G, and A79G retain significant catalytic activity but do not alter the specificity for the N(-1) nucleobase. Effects of nucleobase analog substitutions at N(-1) indicate that U is preferred due to the ability to donate an H-bond in the Watson-Crick face and avoid minor groove steric clash. The results provide information essential for evaluating models of the HDV active site and illustrate multiple substrate kinetic analyses as a practical approach for characterizing structure-function relationships in RNA reactions.
Keywords: Enzyme kinetics; Internal competition; Ribozyme.
Published by Elsevier Inc.
Figures

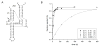



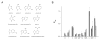

References
-
- Jencks WP. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. - PubMed
-
- Riccitelli N, Lupták A. Chapter Four - HDV Family of Self-Cleaving Ribozymes. In: Garrett AS, editor. Progress in molecular biology and translational science. Academic Press; 2013. pp. 123–171. - PubMed
-
- Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

